A General-Purpose Self-Supervised Model for Computational Pathology
Aug 29, 2023Richard J. Chen, Tong Ding, Ming Y. Lu, Drew F. K. Williamson, Guillaume Jaume, Bowen Chen, Andrew Zhang, Daniel Shao, Andrew H. Song, Muhammad Shaban, Mane Williams, Anurag Vaidya, Sharifa Sahai, Lukas Oldenburg, Luca L. Weishaupt, Judy J. Wang, Walt Williams, Long Phi Le, Georg Gerber, Faisal Mahmood
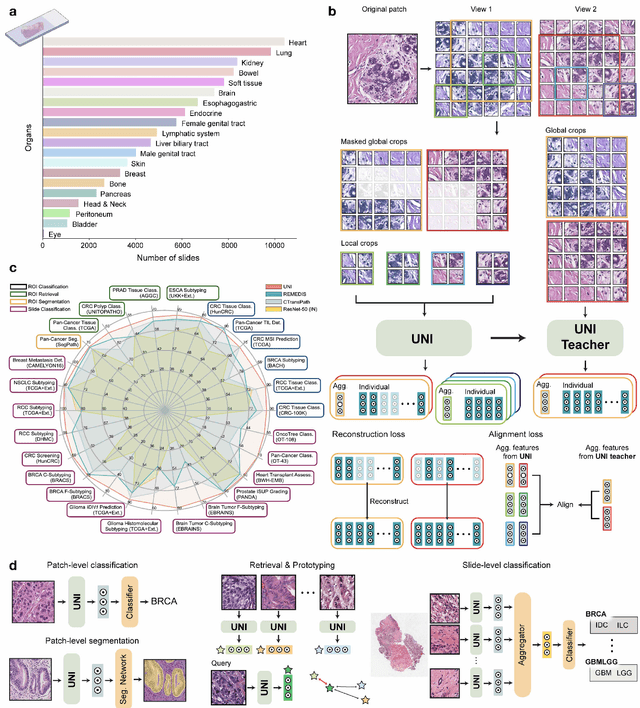
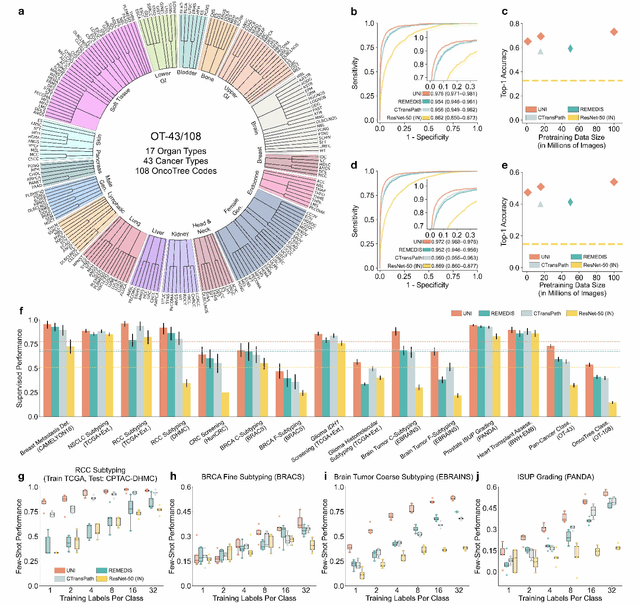
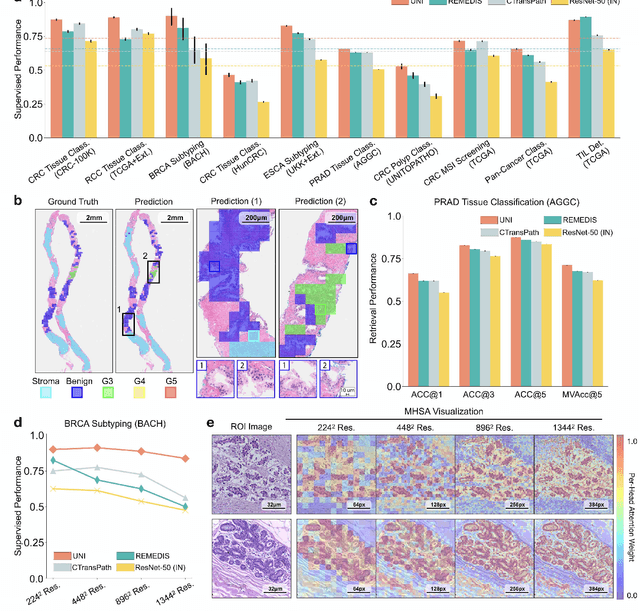

Tissue phenotyping is a fundamental computational pathology (CPath) task in learning objective characterizations of histopathologic biomarkers in anatomic pathology. However, whole-slide imaging (WSI) poses a complex computer vision problem in which the large-scale image resolutions of WSIs and the enormous diversity of morphological phenotypes preclude large-scale data annotation. Current efforts have proposed using pretrained image encoders with either transfer learning from natural image datasets or self-supervised pretraining on publicly-available histopathology datasets, but have not been extensively developed and evaluated across diverse tissue types at scale. We introduce UNI, a general-purpose self-supervised model for pathology, pretrained using over 100 million tissue patches from over 100,000 diagnostic haematoxylin and eosin-stained WSIs across 20 major tissue types, and evaluated on 33 representative CPath clinical tasks in CPath of varying diagnostic difficulties. In addition to outperforming previous state-of-the-art models, we demonstrate new modeling capabilities in CPath such as resolution-agnostic tissue classification, slide classification using few-shot class prototypes, and disease subtyping generalization in classifying up to 108 cancer types in the OncoTree code classification system. UNI advances unsupervised representation learning at scale in CPath in terms of both pretraining data and downstream evaluation, enabling data-efficient AI models that can generalize and transfer to a gamut of diagnostically-challenging tasks and clinical workflows in anatomic pathology.
Weakly Supervised AI for Efficient Analysis of 3D Pathology Samples
Jul 27, 2023Andrew H. Song, Mane Williams, Drew F. K. Williamson, Guillaume Jaume, Andrew Zhang, Bowen Chen, Robert Serafin, Jonathan T. C. Liu, Alex Baras, Anil V. Parwani, Faisal Mahmood
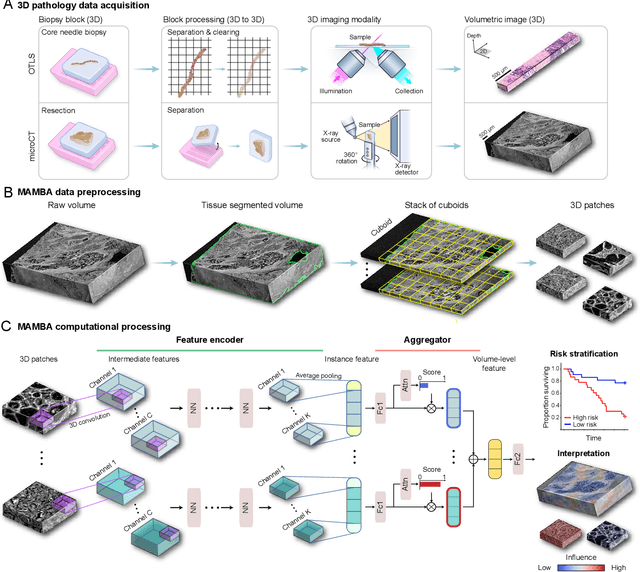
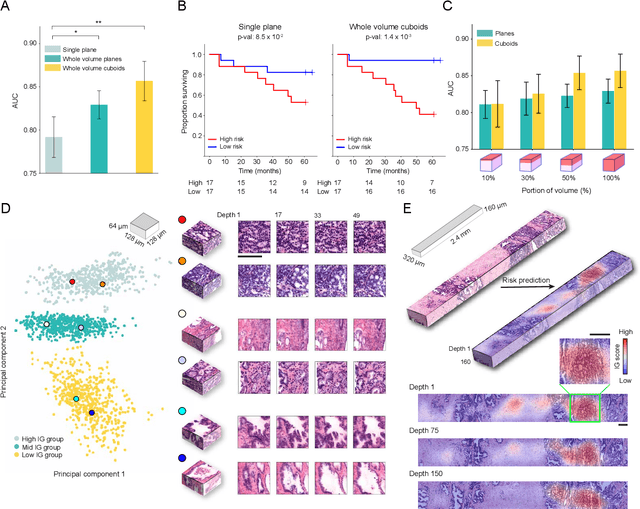
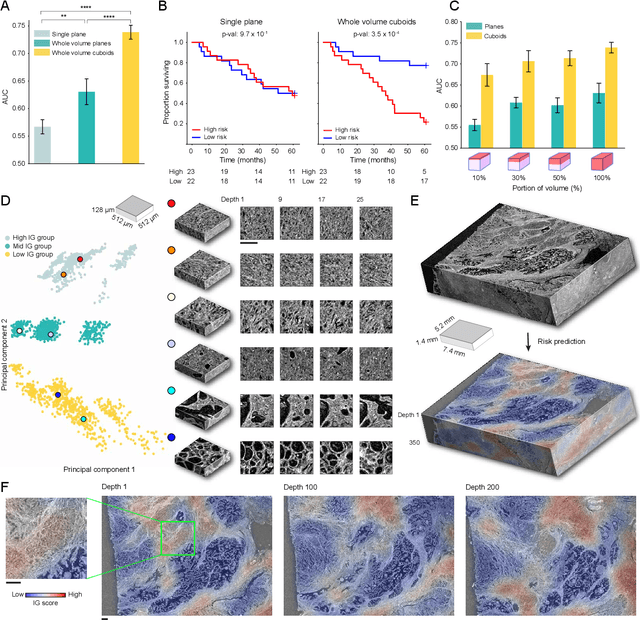
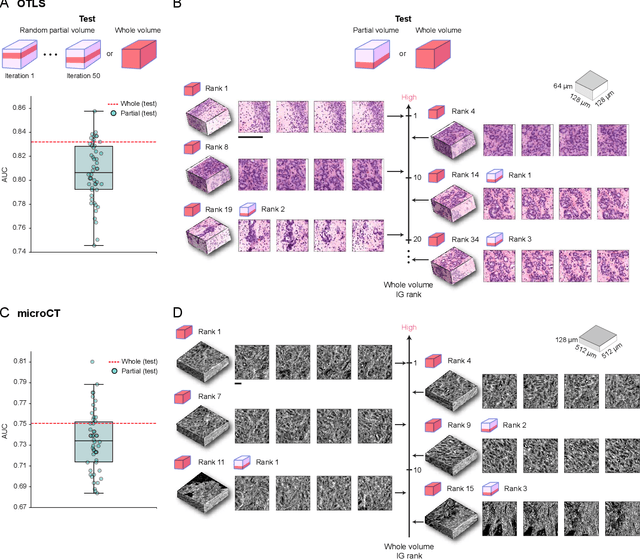
Human tissue and its constituent cells form a microenvironment that is fundamentally three-dimensional (3D). However, the standard-of-care in pathologic diagnosis involves selecting a few two-dimensional (2D) sections for microscopic evaluation, risking sampling bias and misdiagnosis. Diverse methods for capturing 3D tissue morphologies have been developed, but they have yet had little translation to clinical practice; manual and computational evaluations of such large 3D data have so far been impractical and/or unable to provide patient-level clinical insights. Here we present Modality-Agnostic Multiple instance learning for volumetric Block Analysis (MAMBA), a deep-learning-based platform for processing 3D tissue images from diverse imaging modalities and predicting patient outcomes. Archived prostate cancer specimens were imaged with open-top light-sheet microscopy or microcomputed tomography and the resulting 3D datasets were used to train risk-stratification networks based on 5-year biochemical recurrence outcomes via MAMBA. With the 3D block-based approach, MAMBA achieves an area under the receiver operating characteristic curve (AUC) of 0.86 and 0.74, superior to 2D traditional single-slice-based prognostication (AUC of 0.79 and 0.57), suggesting superior prognostication with 3D morphological features. Further analyses reveal that the incorporation of greater tissue volume improves prognostic performance and mitigates risk prediction variability from sampling bias, suggesting the value of capturing larger extents of heterogeneous 3D morphology. With the rapid growth and adoption of 3D spatial biology and pathology techniques by researchers and clinicians, MAMBA provides a general and efficient framework for 3D weakly supervised learning for clinical decision support and can help to reveal novel 3D morphological biomarkers for prognosis and therapeutic response.
Towards a Visual-Language Foundation Model for Computational Pathology
Jul 25, 2023Ming Y. Lu, Bowen Chen, Drew F. K. Williamson, Richard J. Chen, Ivy Liang, Tong Ding, Guillaume Jaume, Igor Odintsov, Andrew Zhang, Long Phi Le, Georg Gerber, Anil V Parwani, Faisal Mahmood
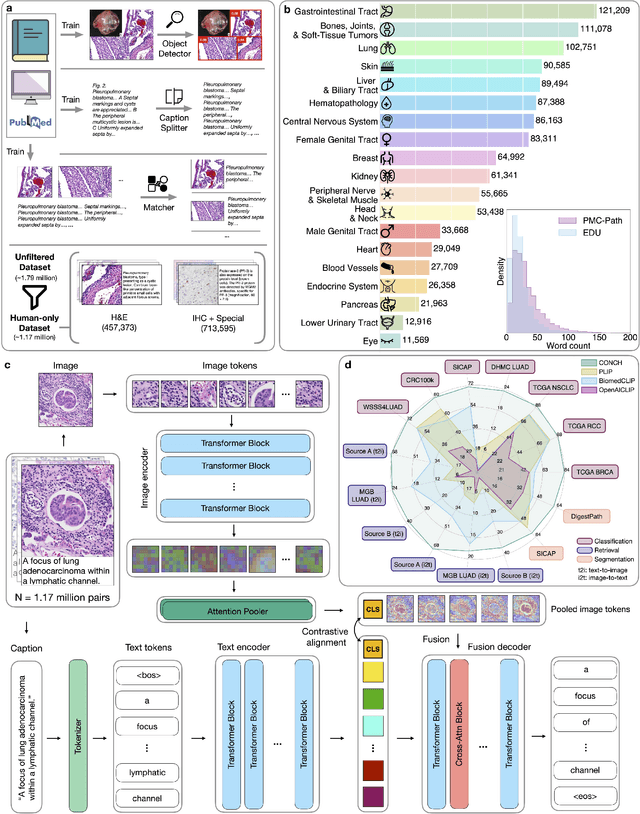
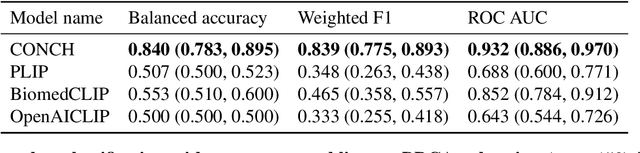
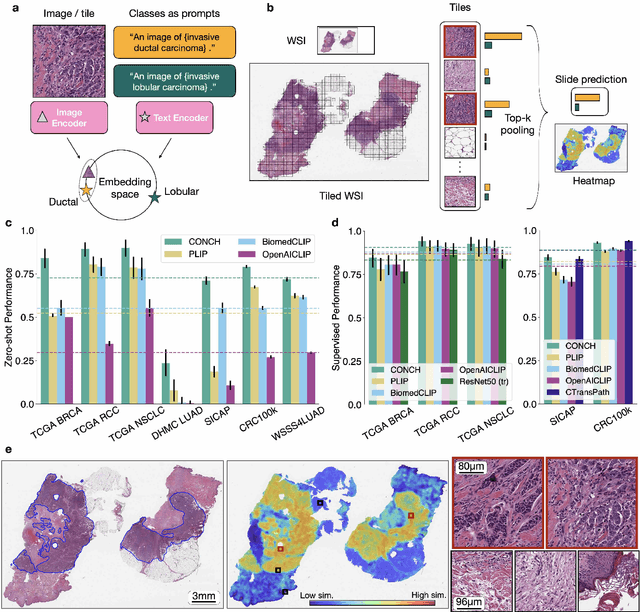
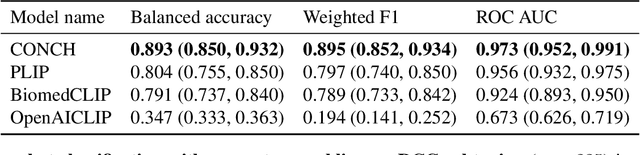
The accelerated adoption of digital pathology and advances in deep learning have enabled the development of powerful models for various pathology tasks across a diverse array of diseases and patient cohorts. However, model training is often difficult due to label scarcity in the medical domain and the model's usage is limited by the specific task and disease for which it is trained. Additionally, most models in histopathology leverage only image data, a stark contrast to how humans teach each other and reason about histopathologic entities. We introduce CONtrastive learning from Captions for Histopathology (CONCH), a visual-language foundation model developed using diverse sources of histopathology images, biomedical text, and notably over 1.17 million image-caption pairs via task-agnostic pretraining. Evaluated on a suite of 13 diverse benchmarks, CONCH can be transferred to a wide range of downstream tasks involving either or both histopathology images and text, achieving state-of-the-art performance on histology image classification, segmentation, captioning, text-to-image and image-to-text retrieval. CONCH represents a substantial leap over concurrent visual-language pretrained systems for histopathology, with the potential to directly facilitate a wide array of machine learning-based workflows requiring minimal or no further supervised fine-tuning.
Modeling Dense Multimodal Interactions Between Biological Pathways and Histology for Survival Prediction
Apr 13, 2023Guillaume Jaume, Anurag Vaidya, Richard Chen, Drew Williamson, Paul Liang, Faisal Mahmood




Integrating whole-slide images (WSIs) and bulk transcriptomics for predicting patient survival can improve our understanding of patient prognosis. However, this multimodal task is particularly challenging due to the different nature of these data: WSIs represent a very high-dimensional spatial description of a tumor, while bulk transcriptomics represent a global description of gene expression levels within that tumor. In this context, our work aims to address two key challenges: (1) how can we tokenize transcriptomics in a semantically meaningful and interpretable way?, and (2) how can we capture dense multimodal interactions between these two modalities? Specifically, we propose to learn biological pathway tokens from transcriptomics that can encode specific cellular functions. Together with histology patch tokens that encode the different morphological patterns in the WSI, we argue that they form appropriate reasoning units for downstream interpretability analyses. We propose fusing both modalities using a memory-efficient multimodal Transformer that can model interactions between pathway and histology patch tokens. Our proposed model, SURVPATH, achieves state-of-the-art performance when evaluated against both unimodal and multimodal baselines on five datasets from The Cancer Genome Atlas. Our interpretability framework identifies key multimodal prognostic factors, and, as such, can provide valuable insights into the interaction between genotype and phenotype, enabling a deeper understanding of the underlying biological mechanisms at play. We make our code public at: https://github.com/ajv012/SurvPath.
Weakly Supervised Joint Whole-Slide Segmentation and Classification in Prostate Cancer
Jan 07, 2023Pushpak Pati, Guillaume Jaume, Zeineb Ayadi, Kevin Thandiackal, Behzad Bozorgtabar, Maria Gabrani, Orcun Goksel

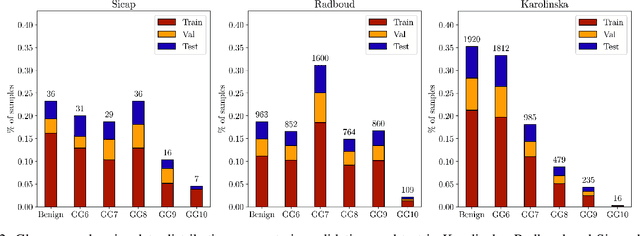
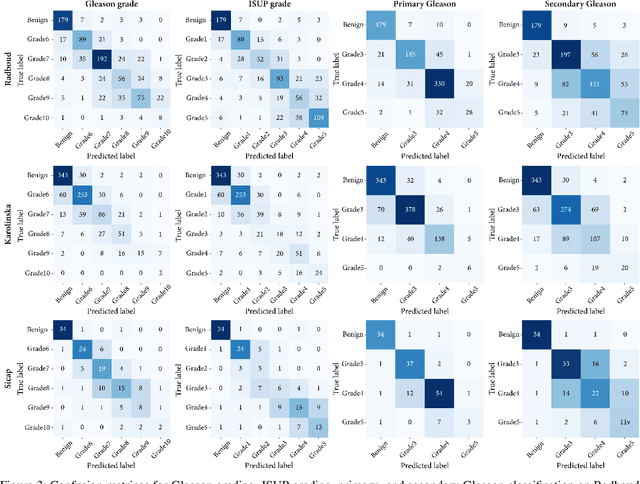
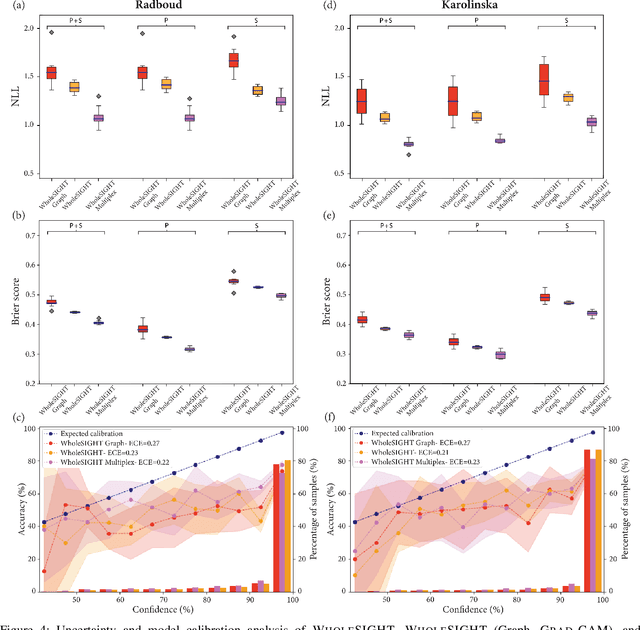
The segmentation and automatic identification of histological regions of diagnostic interest offer a valuable aid to pathologists. However, segmentation methods are hampered by the difficulty of obtaining pixel-level annotations, which are tedious and expensive to obtain for Whole-Slide images (WSI). To remedy this, weakly supervised methods have been developed to exploit the annotations directly available at the image level. However, to our knowledge, none of these techniques is adapted to deal with WSIs. In this paper, we propose WholeSIGHT, a weakly-supervised method, to simultaneously segment and classify WSIs of arbitrary shapes and sizes. Formally, WholeSIGHT first constructs a tissue-graph representation of the WSI, where the nodes and edges depict tissue regions and their interactions, respectively. During training, a graph classification head classifies the WSI and produces node-level pseudo labels via post-hoc feature attribution. These pseudo labels are then used to train a node classification head for WSI segmentation. During testing, both heads simultaneously render class prediction and segmentation for an input WSI. We evaluated WholeSIGHT on three public prostate cancer WSI datasets. Our method achieved state-of-the-art weakly-supervised segmentation performance on all datasets while resulting in better or comparable classification with respect to state-of-the-art weakly-supervised WSI classification methods. Additionally, we quantify the generalization capability of our method in terms of segmentation and classification performance, uncertainty estimation, and model calibration.
Embedding Space Augmentation for Weakly Supervised Learning in Whole-Slide Images
Oct 31, 2022Imaad Zaffar, Guillaume Jaume, Nasir Rajpoot, Faisal Mahmood




Multiple Instance Learning (MIL) is a widely employed framework for learning on gigapixel whole-slide images (WSIs) from WSI-level annotations. In most MIL based analytical pipelines for WSI-level analysis, the WSIs are often divided into patches and deep features for patches (i.e., patch embeddings) are extracted prior to training to reduce the overall computational cost and cope with the GPUs' limited RAM. To overcome this limitation, we present EmbAugmenter, a data augmentation generative adversarial network (DA-GAN) that can synthesize data augmentations in the embedding space rather than in the pixel space, thereby significantly reducing the computational requirements. Experiments on the SICAPv2 dataset show that our approach outperforms MIL without augmentation and is on par with traditional patch-level augmentation for MIL training while being substantially faster.
Differentiable Zooming for Multiple Instance Learning on Whole-Slide Images
Apr 30, 2022Kevin Thandiackal, Boqi Chen, Pushpak Pati, Guillaume Jaume, Drew F. K. Williamson, Maria Gabrani, Orcun Goksel
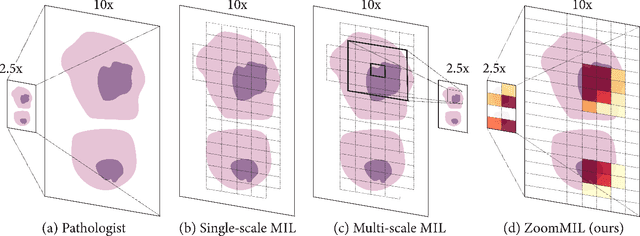
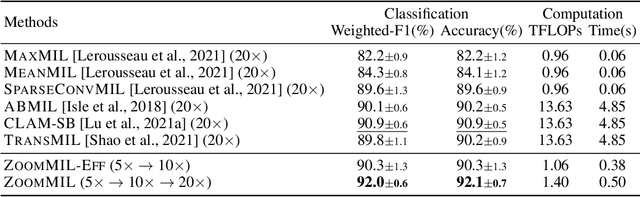
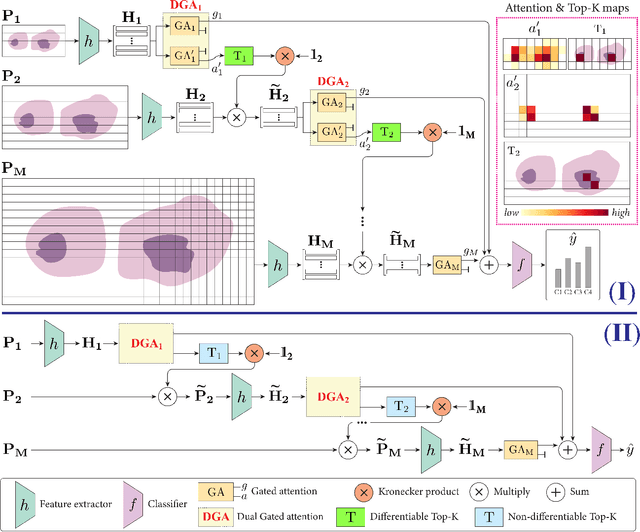
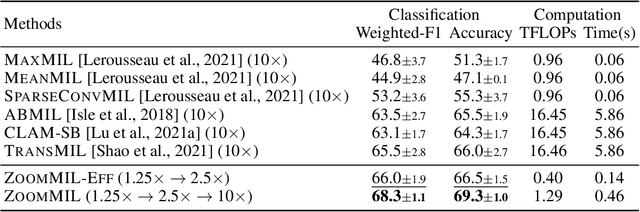
Multiple Instance Learning (MIL) methods have become increasingly popular for classifying giga-pixel sized Whole-Slide Images (WSIs) in digital pathology. Most MIL methods operate at a single WSI magnification, by processing all the tissue patches. Such a formulation induces high computational requirements, and constrains the contextualization of the WSI-level representation to a single scale. A few MIL methods extend to multiple scales, but are computationally more demanding. In this paper, inspired by the pathological diagnostic process, we propose ZoomMIL, a method that learns to perform multi-level zooming in an end-to-end manner. ZoomMIL builds WSI representations by aggregating tissue-context information from multiple magnifications. The proposed method outperforms the state-of-the-art MIL methods in WSI classification on two large datasets, while significantly reducing the computational demands with regard to Floating-Point Operations (FLOPs) and processing time by up to 40x.
BRACS: A Dataset for BReAst Carcinoma Subtyping in H&E Histology Images
Nov 08, 2021Nadia Brancati, Anna Maria Anniciello, Pushpak Pati, Daniel Riccio, Giosuè Scognamiglio, Guillaume Jaume, Giuseppe De Pietro, Maurizio Di Bonito, Antonio Foncubierta, Gerardo Botti, Maria Gabrani, Florinda Feroce, Maria Frucci

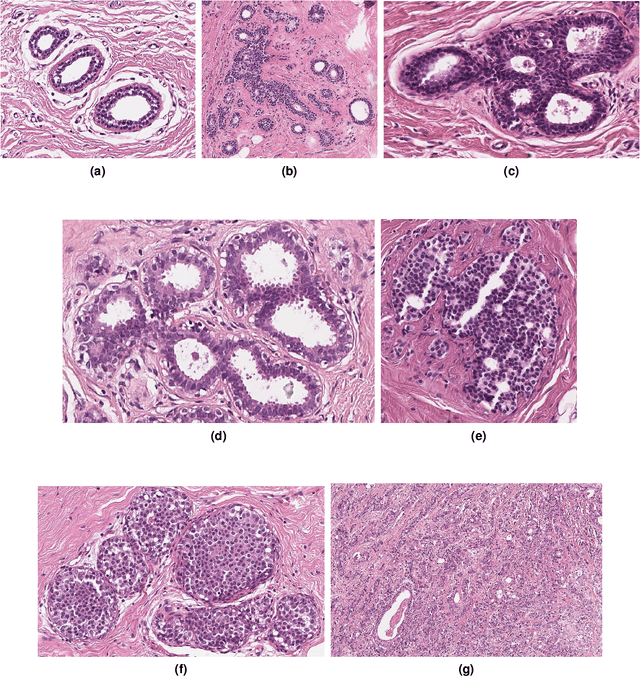
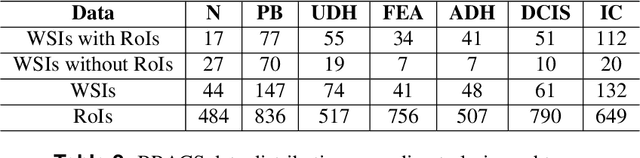
Breast cancer is the most commonly diagnosed cancer and registers the highest number of deaths for women with cancer. Recent advancements in diagnostic activities combined with large-scale screening policies have significantly lowered the mortality rates for breast cancer patients. However, the manual inspection of tissue slides by the pathologists is cumbersome, time-consuming, and is subject to significant inter- and intra-observer variability. Recently, the advent of whole-slide scanning systems have empowered the rapid digitization of pathology slides, and enabled to develop digital workflows. These advances further enable to leverage Artificial Intelligence (AI) to assist, automate, and augment pathological diagnosis. But the AI techniques, especially Deep Learning (DL), require a large amount of high-quality annotated data to learn from. Constructing such task-specific datasets poses several challenges, such as, data-acquisition level constrains, time-consuming and expensive annotations, and anonymization of private information. In this paper, we introduce the BReAst Carcinoma Subtyping (BRACS) dataset, a large cohort of annotated Hematoxylin & Eosin (H&E)-stained images to facilitate the characterization of breast lesions. BRACS contains 547 Whole-Slide Images (WSIs), and 4539 Regions of Interest (ROIs) extracted from the WSIs. Each WSI, and respective ROIs, are annotated by the consensus of three board-certified pathologists into different lesion categories. Specifically, BRACS includes three lesion types, i.e., benign, malignant and atypical, which are further subtyped into seven categories. It is, to the best of our knowledge, the largest annotated dataset for breast cancer subtyping both at WSI- and ROI-level. Further, by including the understudied atypical lesions, BRACS offers an unique opportunity for leveraging AI to better understand their characteristics.
HistoCartography: A Toolkit for Graph Analytics in Digital Pathology
Jul 21, 2021Guillaume Jaume, Pushpak Pati, Valentin Anklin, Antonio Foncubierta, Maria Gabrani




Advances in entity-graph based analysis of histopathology images have brought in a new paradigm to describe tissue composition, and learn the tissue structure-to-function relationship. Entity-graphs offer flexible and scalable representations to characterize tissue organization, while allowing the incorporation of prior pathological knowledge to further support model interpretability and explainability. However, entity-graph analysis requires prerequisites for image-to-graph translation and knowledge of state-of-the-art machine learning algorithms applied to graph-structured data, which can potentially hinder their adoption. In this work, we aim to alleviate these issues by developing HistoCartography, a standardized python API with necessary preprocessing, machine learning and explainability tools to facilitate graph-analytics in computational pathology. Further, we have benchmarked the computational time and performance on multiple datasets across different imaging types and histopathology tasks to highlight the applicability of the API for building computational pathology workflows.
Hierarchical Graph Representations in Digital Pathology
Mar 17, 2021Pushpak Pati, Guillaume Jaume, Antonio Foncubierta, Florinda Feroce, Anna Maria Anniciello, Giosuè Scognamiglio, Nadia Brancati, Maryse Fiche, Estelle Dubruc, Daniel Riccio, Maurizio Di Bonito, Giuseppe De Pietro, Gerardo Botti, Jean-Philippe Thiran, Maria Frucci, Orcun Goksel, Maria Gabrani
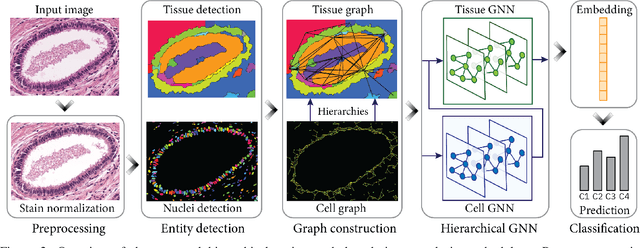
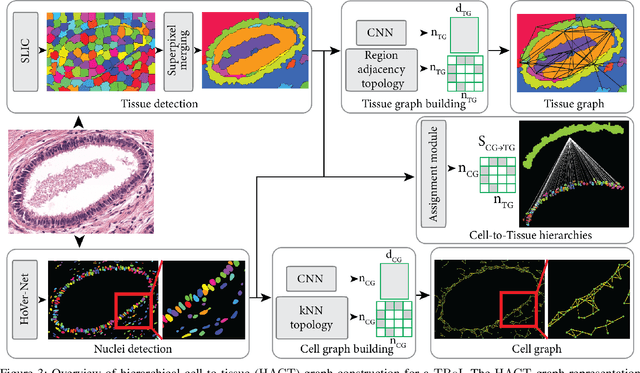
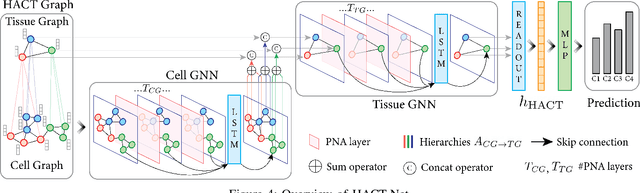
Cancer diagnosis, prognosis, and therapy response predictions from tissue specimens highly depend on the phenotype and topological distribution of constituting histological entities. Thus, adequate tissue representations for encoding histological entities is imperative for computer aided cancer patient care. To this end, several approaches have leveraged cell-graphs that encode cell morphology and organization to denote the tissue information. These allow for utilizing machine learning to map tissue representations to tissue functionality to help quantify their relationship. Though cellular information is crucial, it is incomplete alone to comprehensively characterize complex tissue structure. We herein treat the tissue as a hierarchical composition of multiple types of histological entities from fine to coarse level, capturing multivariate tissue information at multiple levels. We propose a novel multi-level hierarchical entity-graph representation of tissue specimens to model hierarchical compositions that encode histological entities as well as their intra- and inter-entity level interactions. Subsequently, a graph neural network is proposed to operate on the hierarchical entity-graph representation to map the tissue structure to tissue functionality. Specifically, for input histology images we utilize well-defined cells and tissue regions to build HierArchical Cell-to-Tissue (HACT) graph representations, and devise HACT-Net, a graph neural network, to classify such HACT representations. As part of this work, we introduce the BReAst Carcinoma Subtyping (BRACS) dataset, a large cohort of H&E stained breast tumor images, to evaluate our proposed methodology against pathologists and state-of-the-art approaches. Through comparative assessment and ablation studies, our method is demonstrated to yield superior classification results compared to alternative methods as well as pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge