VICE: Visual Identification and Correction of Neural Circuit Errors
May 14, 2021Felix Gonda, Xueying Wang, Johanna Beyer, Markus Hadwiger, Jeff W. Lichtman, Hanspeter Pfister
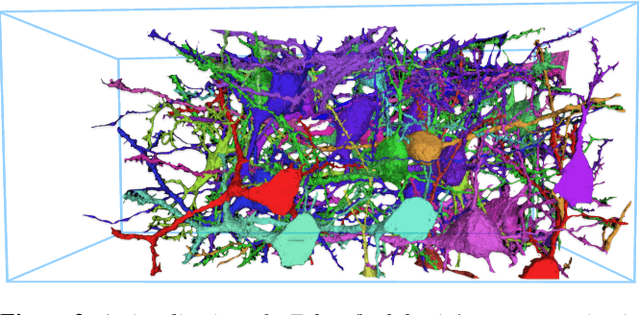

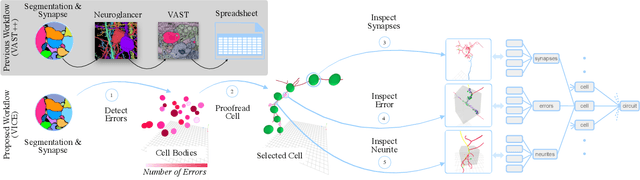
A connectivity graph of neurons at the resolution of single synapses provides scientists with a tool for understanding the nervous system in health and disease. Recent advances in automatic image segmentation and synapse prediction in electron microscopy (EM) datasets of the brain have made reconstructions of neurons possible at the nanometer scale. However, automatic segmentation sometimes struggles to segment large neurons correctly, requiring human effort to proofread its output. General proofreading involves inspecting large volumes to correct segmentation errors at the pixel level, a visually intensive and time-consuming process. This paper presents the design and implementation of an analytics framework that streamlines proofreading, focusing on connectivity-related errors. We accomplish this with automated likely-error detection and synapse clustering that drives the proofreading effort with highly interactive 3D visualizations. In particular, our strategy centers on proofreading the local circuit of a single cell to ensure a basic level of completeness. We demonstrate our framework's utility with a user study and report quantitative and subjective feedback from our users. Overall, users find the framework more efficient for proofreading, understanding evolving graphs, and sharing error correction strategies.
Learning Guided Electron Microscopy with Active Acquisition
Jan 07, 2021Lu Mi, Hao Wang, Yaron Meirovitch, Richard Schalek, Srinivas C. Turaga, Jeff W. Lichtman, Aravinthan D. T. Samuel, Nir Shavit
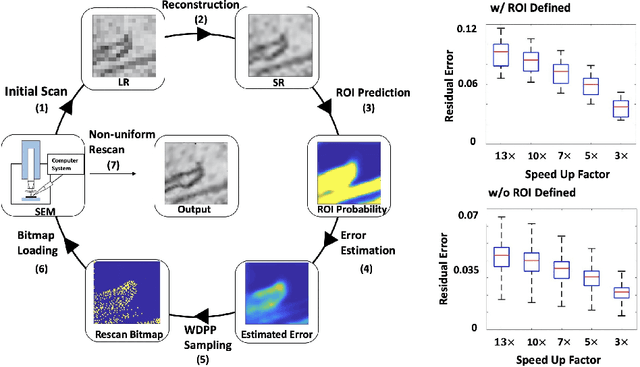

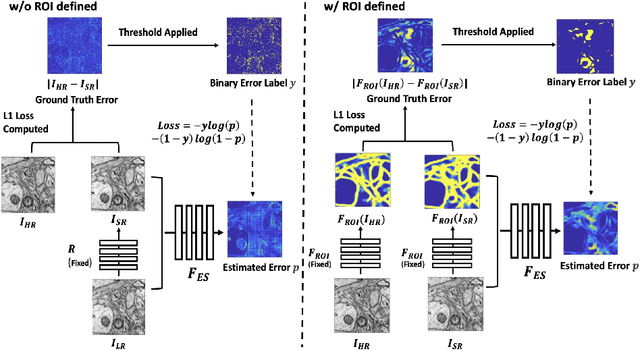
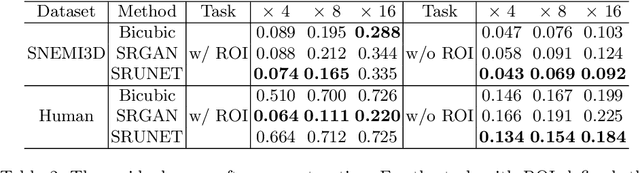
Single-beam scanning electron microscopes (SEM) are widely used to acquire massive data sets for biomedical study, material analysis, and fabrication inspection. Datasets are typically acquired with uniform acquisition: applying the electron beam with the same power and duration to all image pixels, even if there is great variety in the pixels' importance for eventual use. Many SEMs are now able to move the beam to any pixel in the field of view without delay, enabling them, in principle, to invest their time budget more effectively with non-uniform imaging. In this paper, we show how to use deep learning to accelerate and optimize single-beam SEM acquisition of images. Our algorithm rapidly collects an information-lossy image (e.g. low resolution) and then applies a novel learning method to identify a small subset of pixels to be collected at higher resolution based on a trade-off between the saliency and spatial diversity. We demonstrate the efficacy of this novel technique for active acquisition by speeding up the task of collecting connectomic datasets for neurobiology by up to an order of magnitude.
A Topological Nomenclature for 3D Shape Analysis in Connectomics
Sep 27, 2019Abhimanyu Talwar, Zudi Lin, Donglai Wei, Yuesong Wu, Bowen Zheng, Jinglin Zhao, Won-Dong Jang, Xueying Wang, Jeff W. Lichtman, Hanspeter Pfister

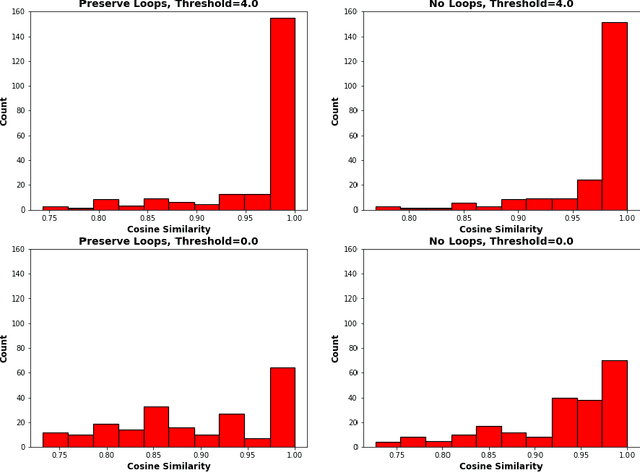
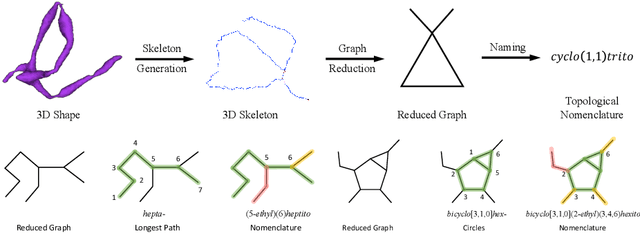
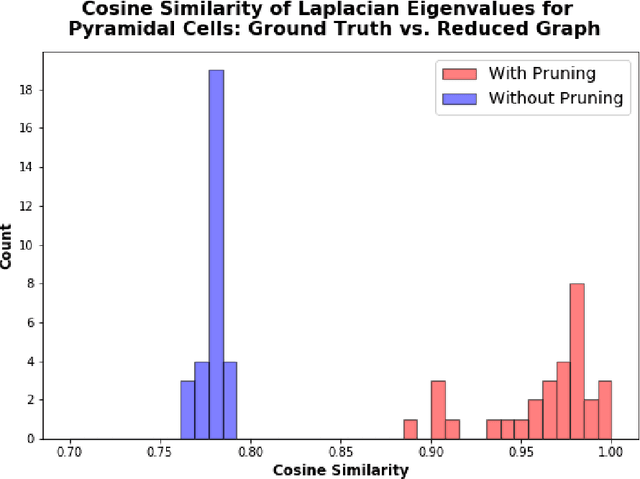
An essential task in nano-scale connectomics is the morphology analysis of neurons and organelles like mitochondria to shed light on their biological properties. However, these biological objects often have tangled parts or complex branching patterns, which makes it hard to abstract, categorize, and manipulate their morphology. Here we propose a topological nomenclature to name these objects like chemical compounds for neuroscience analysis. To this end, we convert the volumetric representation into the topology-preserving reduced graph, develop nomenclature rules for pyramidal neurons and mitochondria from the reduced graph, and learn the feature embedding for shape manipulation. In ablation studies, we show that the proposed reduced graph extraction method yield graphs better in accord with the perception of experts. On 3D shape retrieval and decomposition tasks, we show that the encoded topological nomenclature features achieve better results than state-of-the-art shape descriptors. To advance neuroscience, we will release a 3D mesh dataset of mitochondria and pyramidal neurons reconstructed from a 100{\mu}m cube electron microscopy (EM) volume. Code is publicly available at https://github.com/donglaiw/ibexHelper.
Detecting Synapse Location and Connectivity by Signed Proximity Estimation and Pruning with Deep Nets
Oct 25, 2018Toufiq Parag, Daniel Berger, Lee Kamentsky, Benedikt Staffler, Donglai Wei, Moritz Helmstaedter, Jeff W. Lichtman, Hanspeter Pfister

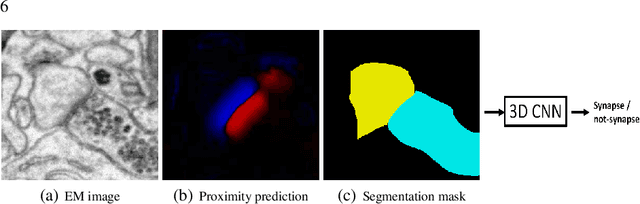
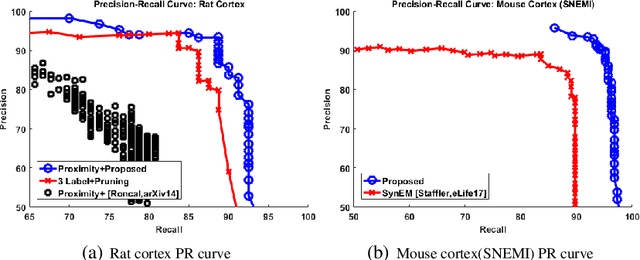
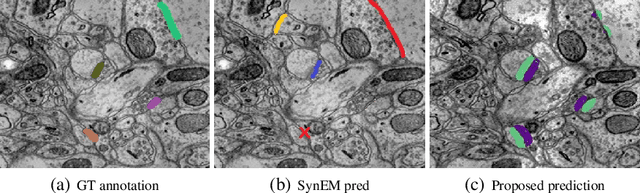
Synaptic connectivity detection is a critical task for neural reconstruction from Electron Microscopy (EM) data. Most of the existing algorithms for synapse detection do not identify the cleft location and direction of connectivity simultaneously. The few methods that computes direction along with contact location have only been demonstrated to work on either dyadic (most common in vertebrate brain) or polyadic (found in fruit fly brain) synapses, but not on both types. In this paper, we present an algorithm to automatically predict the location as well as the direction of both dyadic and polyadic synapses. The proposed algorithm first generates candidate synaptic connections from voxelwise predictions of signed proximity generated by a 3D U-net. A second 3D CNN then prunes the set of candidates to produce the final detection of cleft and connectivity orientation. Experimental results demonstrate that the proposed method outperforms the existing methods for determining synapses in both rodent and fruit fly brain.
Anisotropic EM Segmentation by 3D Affinity Learning and Agglomeration
Aug 03, 2018Toufiq Parag, Fabian Tschopp, William Grisaitis, Srinivas C Turaga, Xuewen Zhang, Brian Matejek, Lee Kamentsky, Jeff W. Lichtman, Hanspeter Pfister
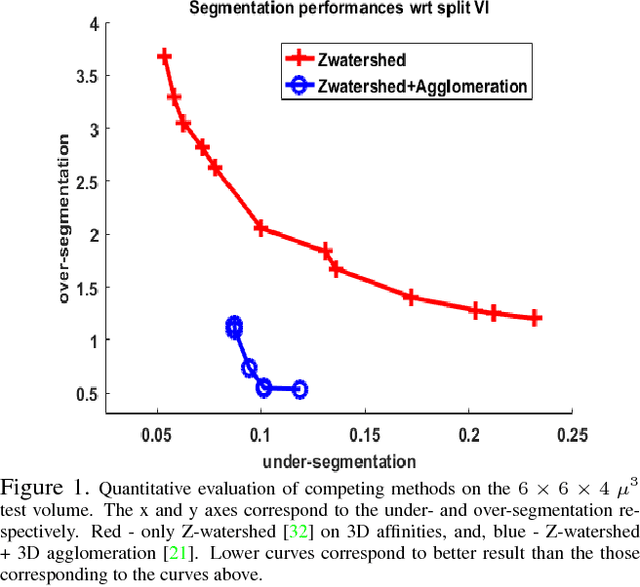
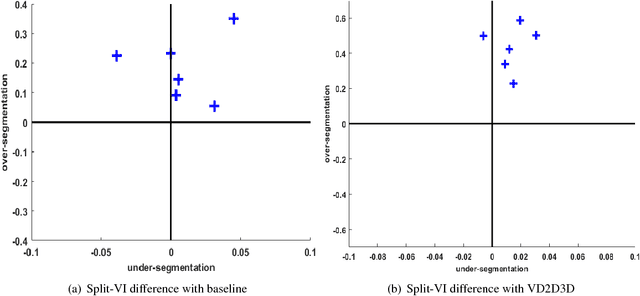
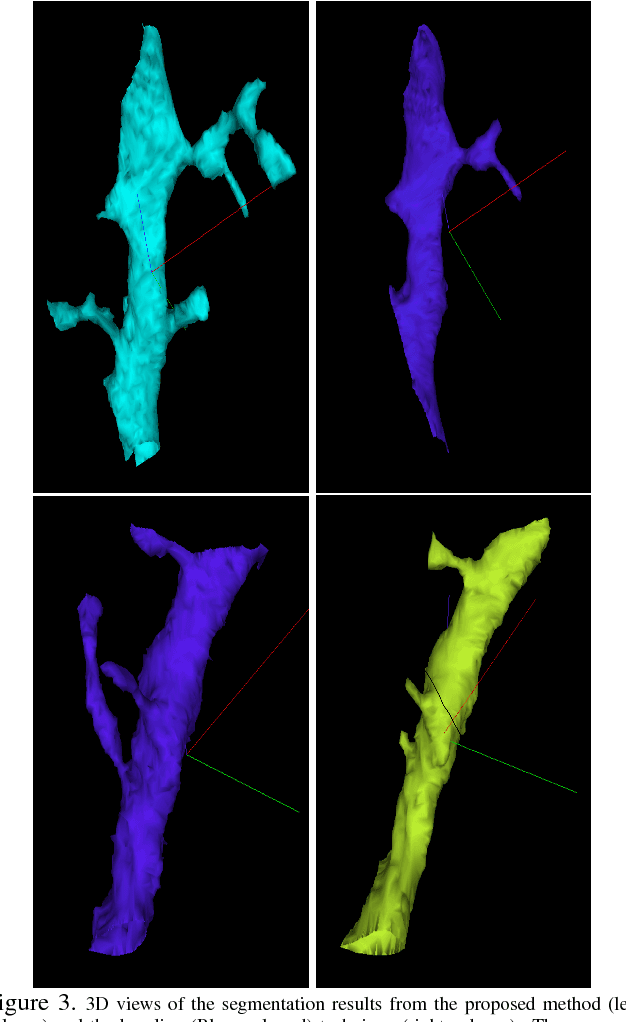
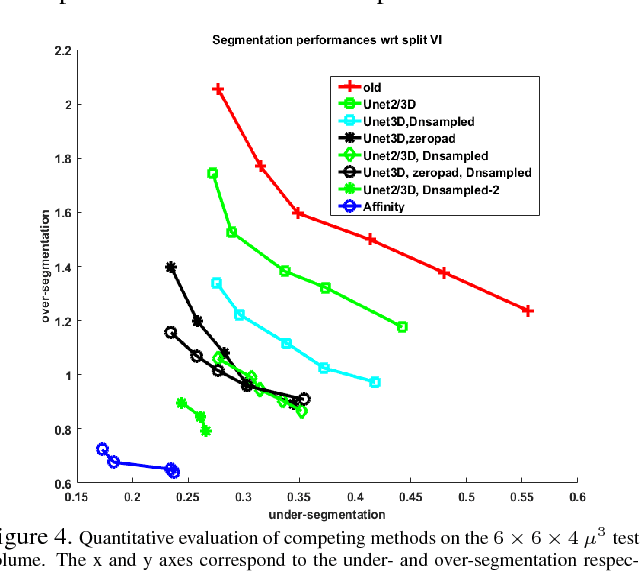
The field of connectomics has recently produced neuron wiring diagrams from relatively large brain regions from multiple animals. Most of these neural reconstructions were computed from isotropic (e.g., FIBSEM) or near isotropic (e.g., SBEM) data. In spite of the remarkable progress on algorithms in recent years, automatic dense reconstruction from anisotropic data remains a challenge for the connectomics community. One significant hurdle in the segmentation of anisotropic data is the difficulty in generating a suitable initial over-segmentation. In this study, we present a segmentation method for anisotropic EM data that agglomerates a 3D over-segmentation computed from the 3D affinity prediction. A 3D U-net is trained to predict 3D affinities by the MALIS approach. Experiments on multiple datasets demonstrates the strength and robustness of the proposed method for anisotropic EM segmentation.
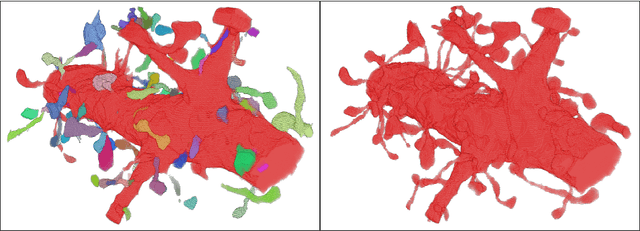
Morphological Error Detection in 3D Segmentations
May 30, 2017David Rolnick, Yaron Meirovitch, Toufiq Parag, Hanspeter Pfister, Viren Jain, Jeff W. Lichtman, Edward S. Boyden, Nir Shavit


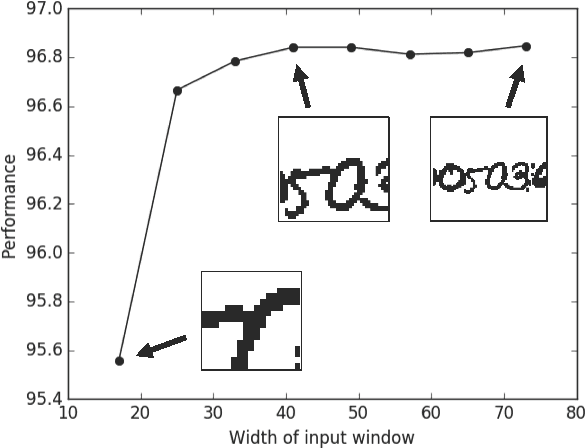
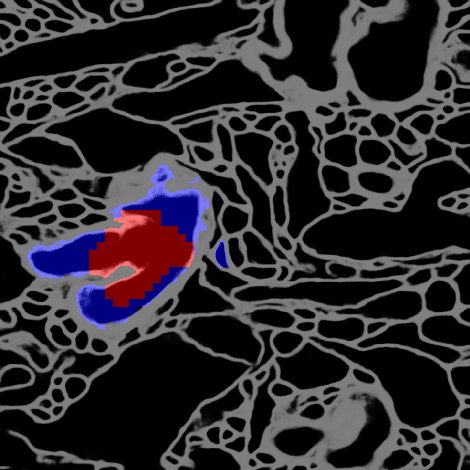
Deep learning algorithms for connectomics rely upon localized classification, rather than overall morphology. This leads to a high incidence of erroneously merged objects. Humans, by contrast, can easily detect such errors by acquiring intuition for the correct morphology of objects. Biological neurons have complicated and variable shapes, which are challenging to learn, and merge errors take a multitude of different forms. We present an algorithm, MergeNet, that shows 3D ConvNets can, in fact, detect merge errors from high-level neuronal morphology. MergeNet follows unsupervised training and operates across datasets. We demonstrate the performance of MergeNet both on a variety of connectomics data and on a dataset created from merged MNIST images.
Guided Proofreading of Automatic Segmentations for Connectomics
Apr 04, 2017Daniel Haehn, Verena Kaynig, James Tompkin, Jeff W. Lichtman, Hanspeter Pfister
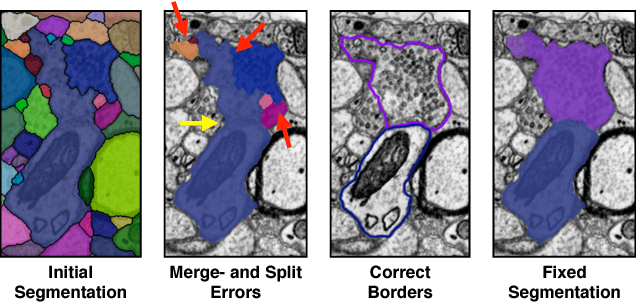
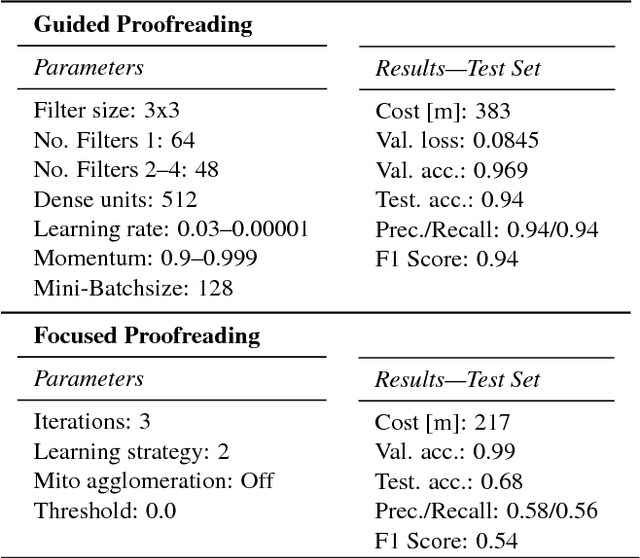
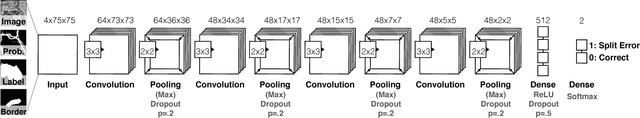
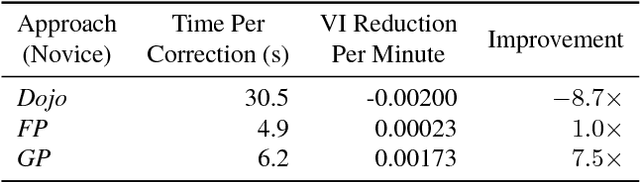
Automatic cell image segmentation methods in connectomics produce merge and split errors, which require correction through proofreading. Previous research has identified the visual search for these errors as the bottleneck in interactive proofreading. To aid error correction, we develop two classifiers that automatically recommend candidate merges and splits to the user. These classifiers use a convolutional neural network (CNN) that has been trained with errors in automatic segmentations against expert-labeled ground truth. Our classifiers detect potentially-erroneous regions by considering a large context region around a segmentation boundary. Corrections can then be performed by a user with yes/no decisions, which reduces variation of information 7.5x faster than previous proofreading methods. We also present a fully-automatic mode that uses a probability threshold to make merge/split decisions. Extensive experiments using the automatic approach and comparing performance of novice and expert users demonstrate that our method performs favorably against state-of-the-art proofreading methods on different connectomics datasets.
Registering large volume serial-section electron microscopy image sets for neural circuit reconstruction using FFT signal whitening
Dec 14, 2016Arthur W. Wetzel, Jennifer Bakal, Markus Dittrich, David G. C. Hildebrand, Josh L. Morgan, Jeff W. Lichtman
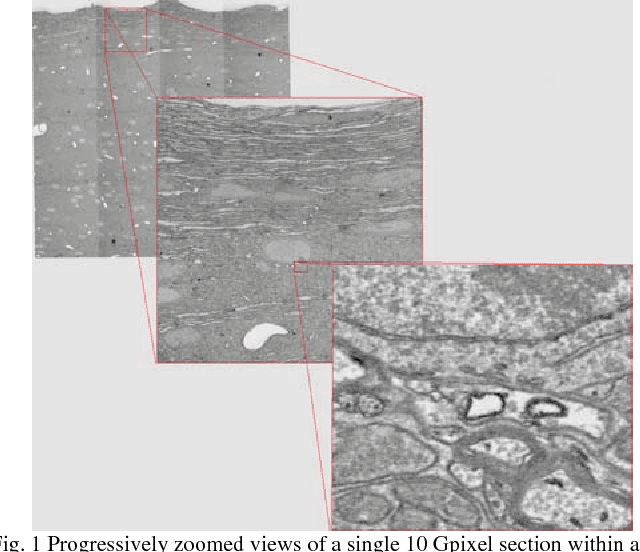
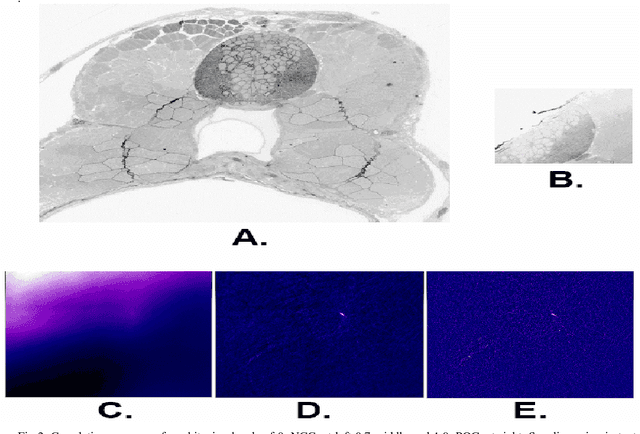
The detailed reconstruction of neural anatomy for connectomics studies requires a combination of resolution and large three-dimensional data capture provided by serial section electron microscopy (ssEM). The convergence of high throughput ssEM imaging and improved tissue preparation methods now allows ssEM capture of complete specimen volumes up to cubic millimeter scale. The resulting multi-terabyte image sets span thousands of serial sections and must be precisely registered into coherent volumetric forms in which neural circuits can be traced and segmented. This paper introduces a Signal Whitening Fourier Transform Image Registration approach (SWiFT-IR) under development at the Pittsburgh Supercomputing Center and its use to align mouse and zebrafish brain datasets acquired using the wafer mapper ssEM imaging technology recently developed at Harvard University. Unlike other methods now used for ssEM registration, SWiFT-IR modifies its spatial frequency response during image matching to maximize a signal-to-noise measure used as its primary indicator of alignment quality. This alignment signal is more robust to rapid variations in biological content and unavoidable data distortions than either phase-only or standard Pearson correlation, thus allowing more precise alignment and statistical confidence. These improvements in turn enable an iterative registration procedure based on projections through multiple sections rather than more typical adjacent-pair matching methods. This projection approach, when coupled with known anatomical constraints and iteratively applied in a multi-resolution pyramid fashion, drives the alignment into a smooth form that properly represents complex and widely varying anatomical content such as the full cross-section zebrafish data.
RhoanaNet Pipeline: Dense Automatic Neural Annotation
Nov 21, 2016Seymour Knowles-Barley, Verena Kaynig, Thouis Ray Jones, Alyssa Wilson, Joshua Morgan, Dongil Lee, Daniel Berger, Narayanan Kasthuri, Jeff W. Lichtman, Hanspeter Pfister
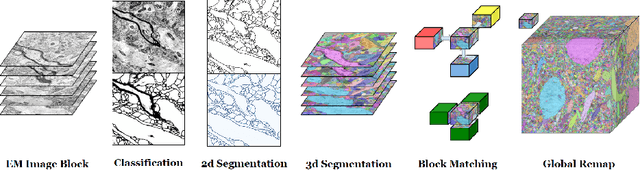
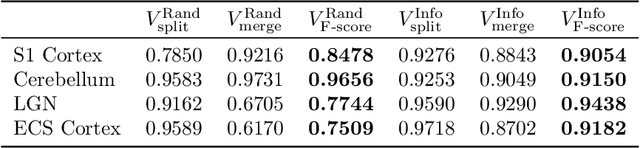
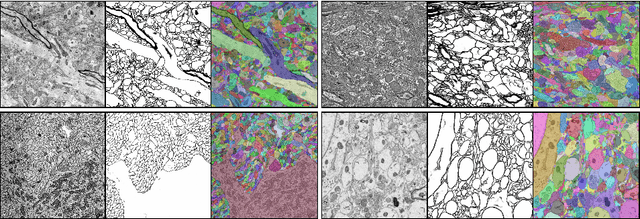
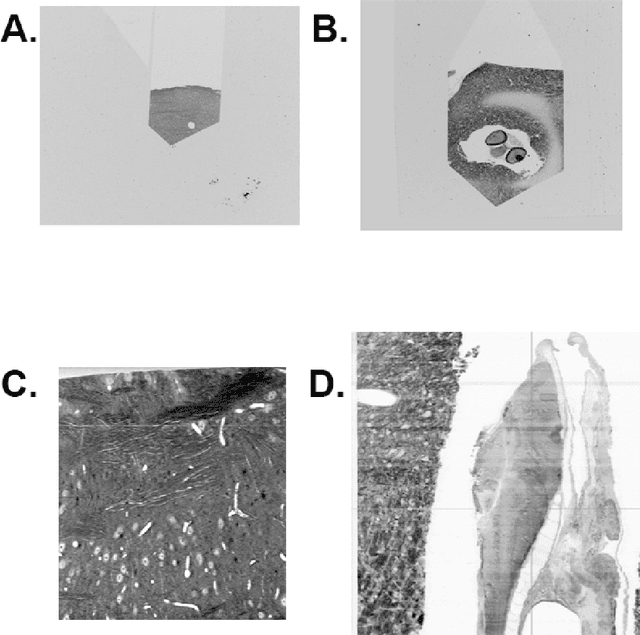
Reconstructing a synaptic wiring diagram, or connectome, from electron microscopy (EM) images of brain tissue currently requires many hours of manual annotation or proofreading (Kasthuri and Lichtman, 2010; Lichtman and Sanes, 2008; Seung, 2009). The desire to reconstruct ever larger and more complex networks has pushed the collection of ever larger EM datasets. A cubic millimeter of raw imaging data would take up 1 PB of storage and present an annotation project that would be impractical without relying heavily on automatic segmentation methods. The RhoanaNet image processing pipeline was developed to automatically segment large volumes of EM data and ease the burden of manual proofreading and annotation. Based on (Kaynig et al., 2015), we updated every stage of the software pipeline to provide better throughput performance and higher quality segmentation results. We used state of the art deep learning techniques to generate improved membrane probability maps, and Gala (Nunez-Iglesias et al., 2014) was used to agglomerate 2D segments into 3D objects. We applied the RhoanaNet pipeline to four densely annotated EM datasets, two from mouse cortex, one from cerebellum and one from mouse lateral geniculate nucleus (LGN). All training and test data is made available for benchmark comparisons. The best segmentation results obtained gave $V^\text{Info}_\text{F-score}$ scores of 0.9054 and 09182 for the cortex datasets, 0.9438 for LGN, and 0.9150 for Cerebellum. The RhoanaNet pipeline is open source software. All source code, training data, test data, and annotations for all four benchmark datasets are available at www.rhoana.org.
Automatic Annotation of Axoplasmic Reticula in Pursuit of Connectomes using High-Resolution Neural EM Data
Apr 16, 2014Ayushi Sinha, William Gray Roncal, Narayanan Kasthuri, Jeff W. Lichtman, Randal Burns, Michael Kazhdan
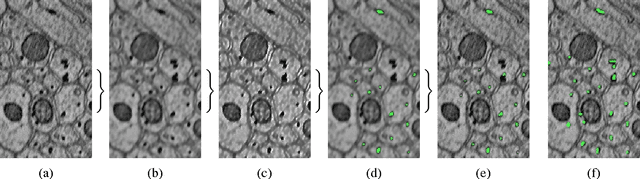

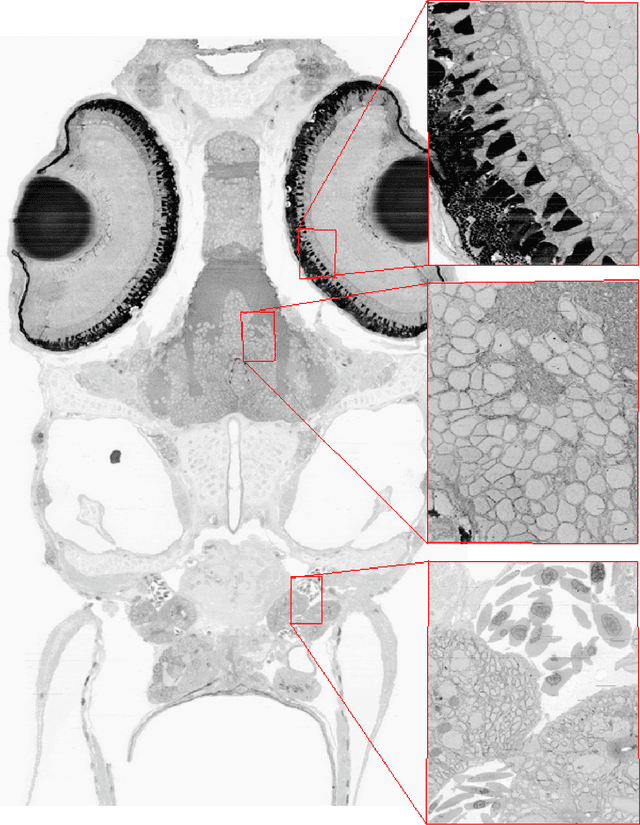
Accurately estimating the wiring diagram of a brain, known as a connectome, at an ultrastructure level is an open research problem. Specifically, precisely tracking neural processes is difficult, especially across many image slices. Here, we propose a novel method to automatically identify and annotate small subcellular structures present in axons, known as axoplasmic reticula, through a 3D volume of high-resolution neural electron microscopy data. Our method produces high precision annotations, which can help improve automatic segmentation by using our results as seeds for segmentation, and as cues to aid segment merging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge