Deep Learning-based Prediction of Breast Cancer Tumor and Immune Phenotypes from Histopathology
Apr 25, 2024Tiago Gonçalves, Dagoberto Pulido-Arias, Julian Willett, Katharina V. Hoebel, Mason Cleveland, Syed Rakin Ahmed, Elizabeth Gerstner, Jayashree Kalpathy-Cramer, Jaime S. Cardoso, Christopher P. Bridge, Albert E. Kim
The interactions between tumor cells and the tumor microenvironment (TME) dictate therapeutic efficacy of radiation and many systemic therapies in breast cancer. However, to date, there is not a widely available method to reproducibly measure tumor and immune phenotypes for each patient's tumor. Given this unmet clinical need, we applied multiple instance learning (MIL) algorithms to assess activity of ten biologically relevant pathways from the hematoxylin and eosin (H&E) slide of primary breast tumors. We employed different feature extraction approaches and state-of-the-art model architectures. Using binary classification, our models attained area under the receiver operating characteristic (AUROC) scores above 0.70 for nearly all gene expression pathways and on some cases, exceeded 0.80. Attention maps suggest that our trained models recognize biologically relevant spatial patterns of cell sub-populations from H&E. These efforts represent a first step towards developing computational H&E biomarkers that reflect facets of the TME and hold promise for augmenting precision oncology.
Automatic classification of prostate MR series type using image content and metadata
Apr 16, 2024Deepa Krishnaswamy, Bálint Kovács, Stefan Denner, Steve Pieper, David Clunie, Christopher P. Bridge, Tina Kapur, Klaus H. Maier-Hein, Andrey Fedorov
With the wealth of medical image data, efficient curation is essential. Assigning the sequence type to magnetic resonance images is necessary for scientific studies and artificial intelligence-based analysis. However, incomplete or missing metadata prevents effective automation. We therefore propose a deep-learning method for classification of prostate cancer scanning sequences based on a combination of image data and DICOM metadata. We demonstrate superior results compared to metadata or image data alone, and make our code publicly available at https://github.com/deepakri201/DICOMScanClassification.
Is Open-Source There Yet? A Comparative Study on Commercial and Open-Source LLMs in Their Ability to Label Chest X-Ray Reports
Feb 19, 2024Felix J. Dorfner, Liv Jürgensen, Leonhard Donle, Fares Al Mohamad, Tobias R. Bodenmann, Mason C. Cleveland, Felix Busch, Lisa C. Adams, James Sato, Thomas Schultz, Albert E. Kim, Jameson Merkow, Keno K. Bressem, Christopher P. Bridge
Introduction: With the rapid advances in large language models (LLMs), there have been numerous new open source as well as commercial models. While recent publications have explored GPT-4 in its application to extracting information of interest from radiology reports, there has not been a real-world comparison of GPT-4 to different leading open-source models. Materials and Methods: Two different and independent datasets were used. The first dataset consists of 540 chest x-ray reports that were created at the Massachusetts General Hospital between July 2019 and July 2021. The second dataset consists of 500 chest x-ray reports from the ImaGenome dataset. We then compared the commercial models GPT-3.5 Turbo and GPT-4 from OpenAI to the open-source models Mistral-7B, Mixtral-8x7B, Llama2-13B, Llama2-70B, QWEN1.5-72B and CheXbert and CheXpert-labeler in their ability to accurately label the presence of multiple findings in x-ray text reports using different prompting techniques. Results: On the ImaGenome dataset, the best performing open-source model was Llama2-70B with micro F1-scores of 0.972 and 0.970 for zero- and few-shot prompts, respectively. GPT-4 achieved micro F1-scores of 0.975 and 0.984, respectively. On the institutional dataset, the best performing open-source model was QWEN1.5-72B with micro F1-scores of 0.952 and 0.965 for zero- and few-shot prompting, respectively. GPT-4 achieved micro F1-scores of 0.975 and 0.973, respectively. Conclusion: In this paper, we show that while GPT-4 is superior to open-source models in zero-shot report labeling, the implementation of few-shot prompting can bring open-source models on par with GPT-4. This shows that open-source models could be a performant and privacy preserving alternative to GPT-4 for the task of radiology report classification.
A generalized framework to predict continuous scores from medical ordinal labels
May 30, 2023Katharina V. Hoebel, Andreanne Lemay, John Peter Campbell, Susan Ostmo, Michael F. Chiang, Christopher P. Bridge, Matthew D. Li, Praveer Singh, Aaron S. Coyner, Jayashree Kalpathy-Cramer




Many variables of interest in clinical medicine, like disease severity, are recorded using discrete ordinal categories such as normal/mild/moderate/severe. These labels are used to train and evaluate disease severity prediction models. However, ordinal categories represent a simplification of an underlying continuous severity spectrum. Using continuous scores instead of ordinal categories is more sensitive to detecting small changes in disease severity over time. Here, we present a generalized framework that accurately predicts continuously valued variables using only discrete ordinal labels during model development. We found that for three clinical prediction tasks, models that take the ordinal relationship of the training labels into account outperformed conventional multi-class classification models. Particularly the continuous scores generated by ordinal classification and regression models showed a significantly higher correlation with expert rankings of disease severity and lower mean squared errors compared to the multi-class classification models. Furthermore, the use of MC dropout significantly improved the ability of all evaluated deep learning approaches to predict continuously valued scores that truthfully reflect the underlying continuous target variable. We showed that accurate continuously valued predictions can be generated even if the model development only involves discrete ordinal labels. The novel framework has been validated on three different clinical prediction tasks and has proven to bridge the gap between discrete ordinal labels and the underlying continuously valued variables.
Improving the repeatability of deep learning models with Monte Carlo dropout
Feb 15, 2022Andreanne Lemay, Katharina Hoebel, Christopher P. Bridge, Brian Befano, Silvia De Sanjosé, Diden Egemen, Ana Cecilia Rodriguez, Mark Schiffman, John Peter Campbell, Jayashree Kalpathy-Cramer




The integration of artificial intelligence into clinical workflows requires reliable and robust models. Repeatability is a key attribute of model robustness. Repeatable models output predictions with low variation during independent tests carried out under similar conditions. During model development and evaluation, much attention is given to classification performance while model repeatability is rarely assessed, leading to the development of models that are unusable in clinical practice. In this work, we evaluate the repeatability of four model types (binary classification, multi-class classification, ordinal classification, and regression) on images that were acquired from the same patient during the same visit. We study the performance of binary, multi-class, ordinal, and regression models on four medical image classification tasks from public and private datasets: knee osteoarthritis, cervical cancer screening, breast density estimation, and retinopathy of prematurity. Repeatability is measured and compared on ResNet and DenseNet architectures. Moreover, we assess the impact of sampling Monte Carlo dropout predictions at test time on classification performance and repeatability. Leveraging Monte Carlo predictions significantly increased repeatability for all tasks on the binary, multi-class, and ordinal models leading to an average reduction of the 95\% limits of agreement by 16% points and of the disagreement rate by 7% points. The classification accuracy improved in most settings along with the repeatability. Our results suggest that beyond about 20 Monte Carlo iterations, there is no further gain in repeatability. In addition to the higher test-retest agreement, Monte Carlo predictions were better calibrated which leads to output probabilities reflecting more accurately the true likelihood of being correctly classified.
Monte Carlo dropout increases model repeatability
Nov 12, 2021Andreanne Lemay, Katharina Hoebel, Christopher P. Bridge, Didem Egemen, Ana Cecilia Rodriguez, Mark Schiffman, John Peter Campbell, Jayashree Kalpathy-Cramer




The integration of artificial intelligence into clinical workflows requires reliable and robust models. Among the main features of robustness is repeatability. Much attention is given to classification performance without assessing the model repeatability, leading to the development of models that turn out to be unusable in practice. In this work, we evaluate the repeatability of four model types on images from the same patient that were acquired during the same visit. We study the performance of binary, multi-class, ordinal, and regression models on three medical image analysis tasks: cervical cancer screening, breast density estimation, and retinopathy of prematurity classification. Moreover, we assess the impact of sampling Monte Carlo dropout predictions at test time on classification performance and repeatability. Leveraging Monte Carlo predictions significantly increased repeatability for all tasks on the binary, multi-class, and ordinal models leading to an average reduction of the 95% limits of agreement by 17% points.
Highdicom: A Python library for standardized encoding of image annotations and machine learning model outputs in pathology and radiology
Jun 14, 2021Christopher P. Bridge, Chris Gorman, Steven Pieper, Sean W. Doyle, Jochen K. Lennerz, Jayashree Kalpathy-Cramer, David A. Clunie, Andriy Y. Fedorov, Markus D. Herrmann




Machine learning is revolutionizing image-based diagnostics in pathology and radiology. ML models have shown promising results in research settings, but their lack of interoperability has been a major barrier for clinical integration and evaluation. The DICOM a standard specifies Information Object Definitions and Services for the representation and communication of digital images and related information, including image-derived annotations and analysis results. However, the complexity of the standard represents an obstacle for its adoption in the ML community and creates a need for software libraries and tools that simplify working with data sets in DICOM format. Here we present the highdicom library, which provides a high-level application programming interface for the Python programming language that abstracts low-level details of the standard and enables encoding and decoding of image-derived information in DICOM format in a few lines of Python code. The highdicom library ties into the extensive Python ecosystem for image processing and machine learning. Simultaneously, by simplifying creation and parsing of DICOM-compliant files, highdicom achieves interoperability with the medical imaging systems that hold the data used to train and run ML models, and ultimately communicate and store model outputs for clinical use. We demonstrate through experiments with slide microscopy and computed tomography imaging, that, by bridging these two ecosystems, highdicom enables developers to train and evaluate state-of-the-art ML models in pathology and radiology while remaining compliant with the DICOM standard and interoperable with clinical systems at all stages. To promote standardization of ML research and streamline the ML model development and deployment process, we made the library available free and open-source.
Addressing catastrophic forgetting for medical domain expansion
Mar 24, 2021Sharut Gupta, Praveer Singh, Ken Chang, Liangqiong Qu, Mehak Aggarwal, Nishanth Arun, Ashwin Vaswani, Shruti Raghavan, Vibha Agarwal, Mishka Gidwani, Katharina Hoebel, Jay Patel, Charles Lu, Christopher P. Bridge, Daniel L. Rubin, Jayashree Kalpathy-Cramer

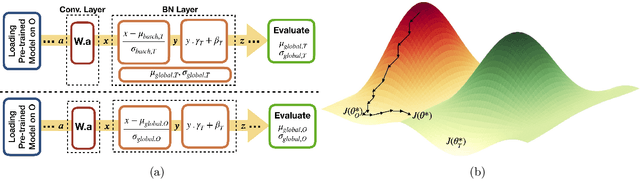
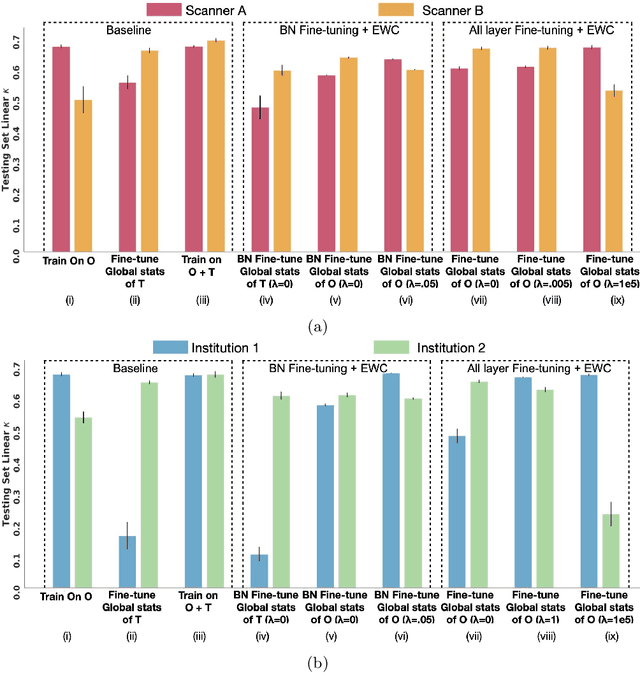
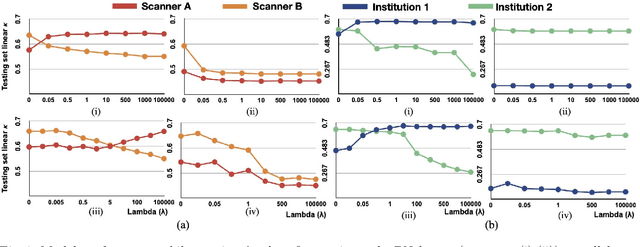
Model brittleness is a key concern when deploying deep learning models in real-world medical settings. A model that has high performance at one institution may suffer a significant decline in performance when tested at other institutions. While pooling datasets from multiple institutions and retraining may provide a straightforward solution, it is often infeasible and may compromise patient privacy. An alternative approach is to fine-tune the model on subsequent institutions after training on the original institution. Notably, this approach degrades model performance at the original institution, a phenomenon known as catastrophic forgetting. In this paper, we develop an approach to address catastrophic forget-ting based on elastic weight consolidation combined with modulation of batch normalization statistics under two scenarios: first, for expanding the domain from one imaging system's data to another imaging system's, and second, for expanding the domain from a large multi-institutional dataset to another single institution dataset. We show that our approach outperforms several other state-of-the-art approaches and provide theoretical justification for the efficacy of batch normalization modulation. The results of this study are generally applicable to the deployment of any clinical deep learning model which requires domain expansion.
"Name that manufacturer". Relating image acquisition bias with task complexity when training deep learning models: experiments on head CT
Aug 19, 2020Giorgio Pietro Biondetti, Romane Gauriau, Christopher P. Bridge, Charles Lu, Katherine P. Andriole




As interest in applying machine learning techniques for medical images continues to grow at a rapid pace, models are starting to be developed and deployed for clinical applications. In the clinical AI model development lifecycle (described by Lu et al. [1]), a crucial phase for machine learning scientists and clinicians is the proper design and collection of the data cohort. The ability to recognize various forms of biases and distribution shifts in the dataset is critical at this step. While it remains difficult to account for all potential sources of bias, techniques can be developed to identify specific types of bias in order to mitigate their impact. In this work we analyze how the distribution of scanner manufacturers in a dataset can contribute to the overall bias of deep learning models. We evaluate convolutional neural networks (CNN) for both classification and segmentation tasks, specifically two state-of-the-art models: ResNet [2] for classification and U-Net [3] for segmentation. We demonstrate that CNNs can learn to distinguish the imaging scanner manufacturer and that this bias can substantially impact model performance for both classification and segmentation tasks. By creating an original synthesis dataset of brain data mimicking the presence of more or less subtle lesions we also show that this bias is related to the difficulty of the task. Recognition of such bias is critical to develop robust, generalizable models that will be crucial for clinical applications in real-world data distributions.
Fully-Automated Analysis of Body Composition from CT in Cancer Patients Using Convolutional Neural Networks
Aug 11, 2018Christopher P. Bridge, Michael Rosenthal, Bradley Wright, Gopal Kotecha, Florian Fintelmann, Fabian Troschel, Nityanand Miskin, Khanant Desai, William Wrobel, Ana Babic, Natalia Khalaf, Lauren Brais, Marisa Welch, Caitlin Zellers, Neil Tenenholtz, Mark Michalski, Brian Wolpin, Katherine Andriole




The amounts of muscle and fat in a person's body, known as body composition, are correlated with cancer risks, cancer survival, and cardiovascular risk. The current gold standard for measuring body composition requires time-consuming manual segmentation of CT images by an expert reader. In this work, we describe a two-step process to fully automate the analysis of CT body composition using a DenseNet to select the CT slice and U-Net to perform segmentation. We train and test our methods on independent cohorts. Our results show Dice scores (0.95-0.98) and correlation coefficients (R=0.99) that are favorable compared to human readers. These results suggest that fully automated body composition analysis is feasible, which could enable both clinical use and large-scale population studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge