Beyond Multiple Instance Learning: Full Resolution All-In-Memory End-To-End Pathology Slide Modeling
Mar 07, 2024Gabriele Campanella, Eugene Fluder, Jennifer Zeng, Chad Vanderbilt, Thomas J. Fuchs


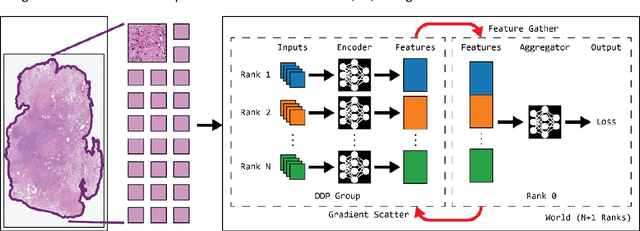

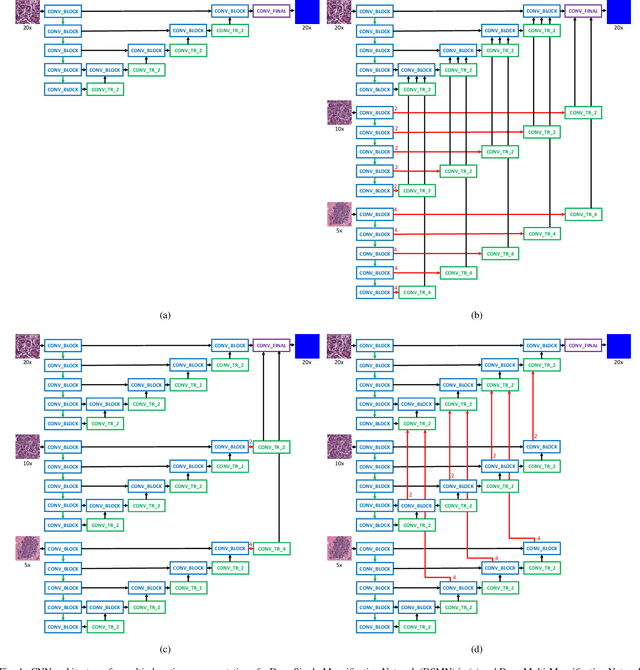
Artificial Intelligence (AI) has great potential to improve health outcomes by training systems on vast digitized clinical datasets. Computational Pathology, with its massive amounts of microscopy image data and impact on diagnostics and biomarkers, is at the forefront of this development. Gigapixel pathology slides pose a unique challenge due to their enormous size and are usually divided into tens of thousands of smaller tiles for analysis. This results in a discontinuity in the machine learning process by separating the training of tile-level encoders from slide-level aggregators and the need to adopt weakly supervised learning strategies. Training models from entire pathology slides end-to-end has been largely unexplored due to its computational challenges. To overcome this problem, we propose a novel approach to jointly train both a tile encoder and a slide-aggregator fully in memory and end-to-end at high-resolution, bridging the gap between input and slide-level supervision. While more computationally expensive, detailed quantitative validation shows promise for large-scale pre-training of pathology foundation models.
Computational Pathology at Health System Scale -- Self-Supervised Foundation Models from Three Billion Images
Oct 10, 2023Gabriele Campanella, Ricky Kwan, Eugene Fluder, Jennifer Zeng, Aryeh Stock, Brandon Veremis, Alexandros D. Polydorides, Cyrus Hedvat, Adam Schoenfeld, Chad Vanderbilt, Patricia Kovatch, Carlos Cordon-Cardo, Thomas J. Fuchs

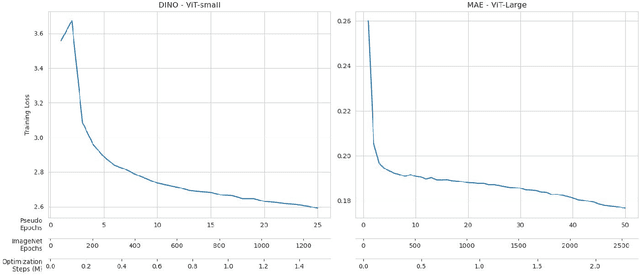
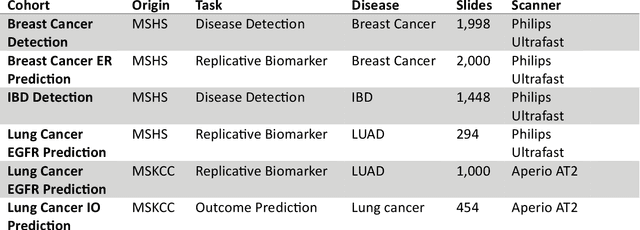
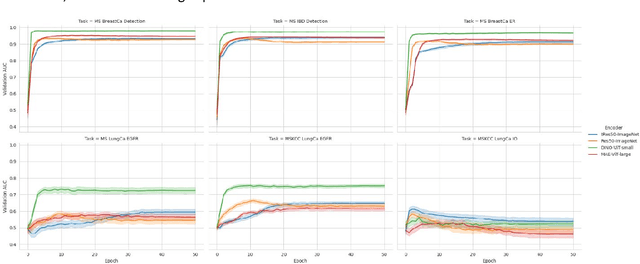
Recent breakthroughs in self-supervised learning have enabled the use of large unlabeled datasets to train visual foundation models that can generalize to a variety of downstream tasks. While this training paradigm is well suited for the medical domain where annotations are scarce, large-scale pre-training in the medical domain, and in particular pathology, has not been extensively studied. Previous work in self-supervised learning in pathology has leveraged smaller datasets for both pre-training and evaluating downstream performance. The aim of this project is to train the largest academic foundation model and benchmark the most prominent self-supervised learning algorithms by pre-training and evaluating downstream performance on large clinical pathology datasets. We collected the largest pathology dataset to date, consisting of over 3 billion images from over 423 thousand microscopy slides. We compared pre-training of visual transformer models using the masked autoencoder (MAE) and DINO algorithms. We evaluated performance on six clinically relevant tasks from three anatomic sites and two institutions: breast cancer detection, inflammatory bowel disease detection, breast cancer estrogen receptor prediction, lung adenocarcinoma EGFR mutation prediction, and lung cancer immunotherapy response prediction. Our results demonstrate that pre-training on pathology data is beneficial for downstream performance compared to pre-training on natural images. Additionally, the DINO algorithm achieved better generalization performance across all tasks tested. The presented results signify a phase change in computational pathology research, paving the way into a new era of more performant models based on large-scale, parallel pre-training at the billion-image scale.
Virchow: A Million-Slide Digital Pathology Foundation Model
Sep 21, 2023Eugene Vorontsov, Alican Bozkurt, Adam Casson, George Shaikovski, Michal Zelechowski, Siqi Liu, Philippe Mathieu, Alexander van Eck, Donghun Lee, Julian Viret, Eric Robert, Yi Kan Wang, Jeremy D. Kunz, Matthew C. H. Lee, Jan Bernhard, Ran A. Godrich, Gerard Oakley, Ewan Millar, Matthew Hanna, Juan Retamero, William A. Moye, Razik Yousfi, Christopher Kanan, David Klimstra, Brandon Rothrock, Thomas J. Fuchs
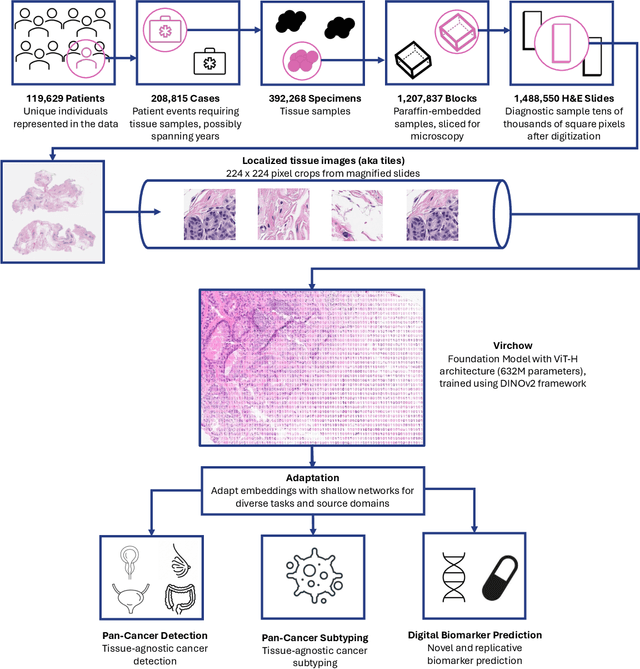
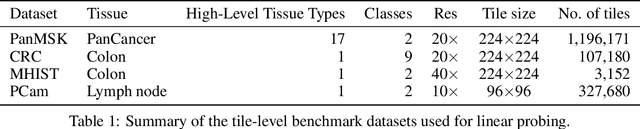
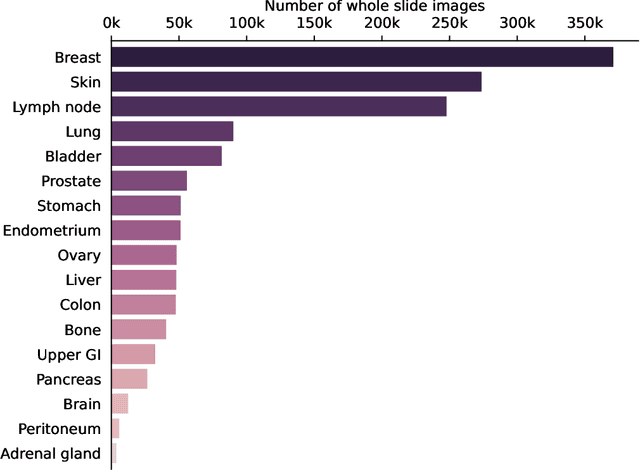
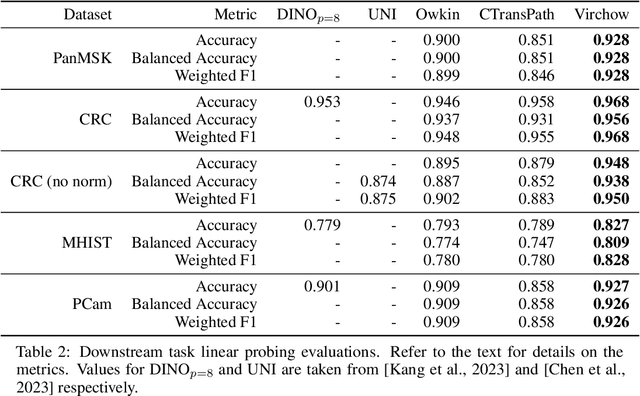
Computational pathology uses artificial intelligence to enable precision medicine and decision support systems through the analysis of whole slide images. It has the potential to revolutionize the diagnosis and treatment of cancer. However, a major challenge to this objective is that for many specific computational pathology tasks the amount of data is inadequate for development. To address this challenge, we created Virchow, a 632 million parameter deep neural network foundation model for computational pathology. Using self-supervised learning, Virchow is trained on 1.5 million hematoxylin and eosin stained whole slide images from diverse tissue groups, which is orders of magnitude more data than previous works. When evaluated on downstream tasks including tile-level pan-cancer detection and subtyping and slide-level biomarker prediction, Virchow outperforms state-of-the-art systems both on internal datasets drawn from the same population as the pretraining data as well as external public datasets. Virchow achieves 93% balanced accuracy for pancancer tile classification, and AUCs of 0.983 for colon microsatellite instability status prediction and 0.967 for breast CDH1 status prediction. The gains in performance highlight the importance of pretraining on massive pathology image datasets, suggesting pretraining on even larger datasets could continue improving performance for many high-impact applications where limited amounts of training data are available, such as drug outcome prediction.
Deep conditional transformation models for survival analysis
Oct 20, 2022Gabriele Campanella, Lucas Kook, Ida Häggström, Torsten Hothorn, Thomas J. Fuchs
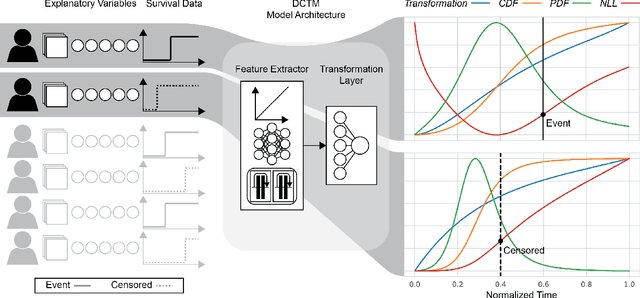
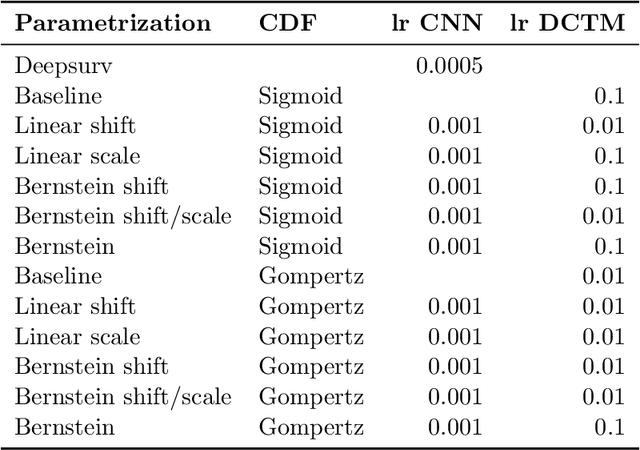
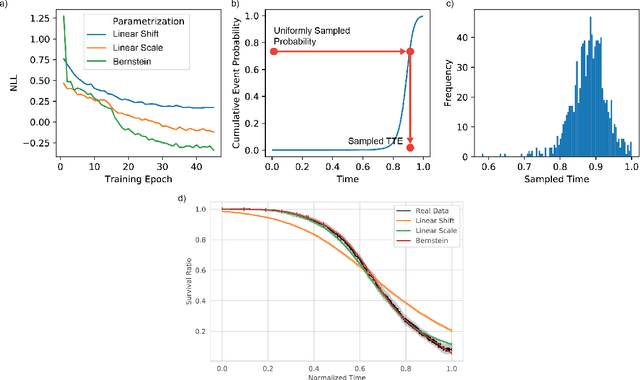
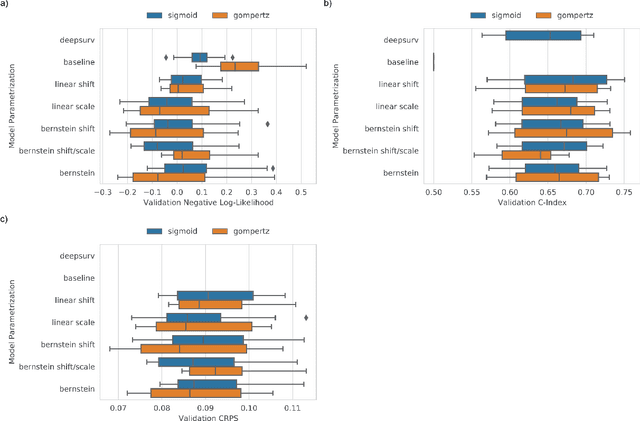
An every increasing number of clinical trials features a time-to-event outcome and records non-tabular patient data, such as magnetic resonance imaging or text data in the form of electronic health records. Recently, several neural-network based solutions have been proposed, some of which are binary classifiers. Parametric, distribution-free approaches which make full use of survival time and censoring status have not received much attention. We present deep conditional transformation models (DCTMs) for survival outcomes as a unifying approach to parametric and semiparametric survival analysis. DCTMs allow the specification of non-linear and non-proportional hazards for both tabular and non-tabular data and extend to all types of censoring and truncation. On real and semi-synthetic data, we show that DCTMs compete with state-of-the-art DL approaches to survival analysis.
Deep Learning-Based Objective and Reproducible Osteosarcoma Chemotherapy Response Assessment and Outcome Prediction
Aug 09, 2022David Joon Ho, Narasimhan P. Agaram, Marc-Henri Jean, Stephanie D. Suser, Cynthia Chu, Chad M. Vanderbilt, Paul A. Meyers, Leonard H. Wexler, John H. Healey, Thomas J. Fuchs, Meera R. Hameed

Osteosarcoma is the most common primary bone cancer whose standard treatment includes pre-operative chemotherapy followed by resection. Chemotherapy response is used for predicting prognosis and further management of patients. Necrosis is routinely assessed post-chemotherapy from histology slides on resection specimens where necrosis ratio is defined as the ratio of necrotic tumor to overall tumor. Patients with necrosis ratio >=90% are known to have better outcome. Manual microscopic review of necrosis ratio from multiple glass slides is semi-quantitative and can have intra- and inter-observer variability. We propose an objective and reproducible deep learning-based approach to estimate necrosis ratio with outcome prediction from scanned hematoxylin and eosin whole slide images. We collected 103 osteosarcoma cases with 3134 WSIs to train our deep learning model, to validate necrosis ratio assessment, and to evaluate outcome prediction. We trained Deep Multi-Magnification Network to segment multiple tissue subtypes including viable tumor and necrotic tumor in pixel-level and to calculate case-level necrosis ratio from multiple WSIs. We showed necrosis ratio estimated by our segmentation model highly correlates with necrosis ratio from pathology reports manually assessed by experts where mean absolute differences for Grades IV (100%), III (>=90%), and II (>=50% and <90%) necrosis response are 4.4%, 4.5%, and 17.8%, respectively. We successfully stratified patients to predict overall survival with p=10^-6 and progression-free survival with p=0.012. Our reproducible approach without variability enabled us to tune cutoff thresholds, specifically for our model and our data set, to 80% for OS and 60% for PFS. Our study indicates deep learning can support pathologists as an objective tool to analyze osteosarcoma from histology for assessing treatment response and predicting patient outcome.
Deep Interactive Learning-based ovarian cancer segmentation of H&E-stained whole slide images to study morphological patterns of BRCA mutation
Mar 28, 2022David Joon Ho, M. Herman Chui, Chad M. Vanderbilt, Jiwon Jung, Mark E. Robson, Chan-Sik Park, Jin Roh, Thomas J. Fuchs
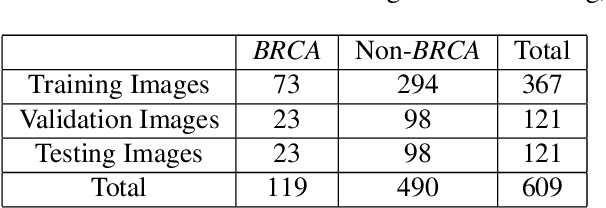
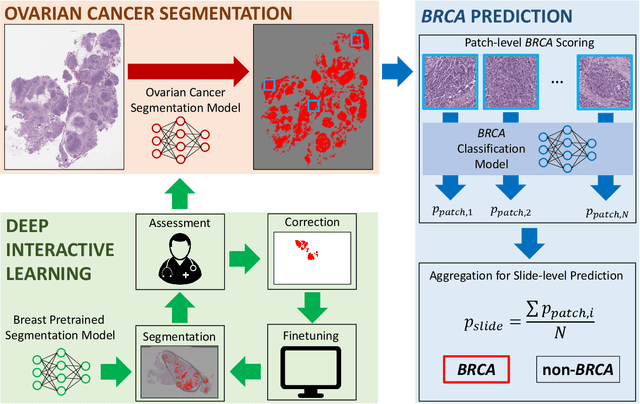
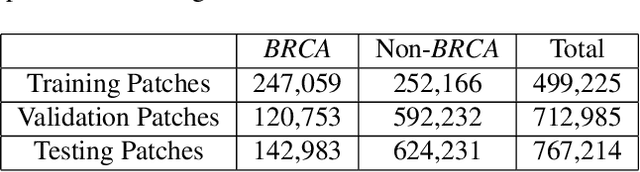

Deep learning has been widely used to analyze digitized hematoxylin and eosin (H&E)-stained histopathology whole slide images. Automated cancer segmentation using deep learning can be used to diagnose malignancy and to find novel morphological patterns to predict molecular subtypes. To train pixel-wise cancer segmentation models, manual annotation from pathologists is generally a bottleneck due to its time-consuming nature. In this paper, we propose Deep Interactive Learning with a pretrained segmentation model from a different cancer type to reduce manual annotation time. Instead of annotating all pixels from cancer and non-cancer regions on giga-pixel whole slide images, an iterative process of annotating mislabeled regions from a segmentation model and training/finetuning the model with the additional annotation can reduce the time. Especially, employing a pretrained segmentation model can further reduce the time than starting annotation from scratch. We trained an accurate ovarian cancer segmentation model with a pretrained breast segmentation model by 3.5 hours of manual annotation which achieved intersection-over-union of 0.74, recall of 0.86, and precision of 0.84. With automatically extracted high-grade serous ovarian cancer patches, we attempted to train another deep learning model to predict BRCA mutation. The segmentation model and code have been released at https://github.com/MSKCC-Computational-Pathology/DMMN-ovary.
EPIC-Survival: End-to-end Part Inferred Clustering for Survival Analysis, Featuring Prognostic Stratification Boosting
Jan 28, 2021Hassan Muhammad, Chensu Xie, Carlie S. Sigel, Michael Doukas, Lindsay Alpert, Thomas J. Fuchs
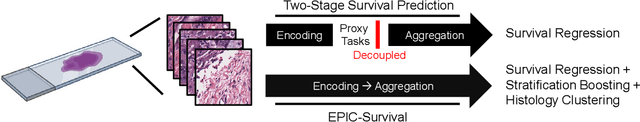

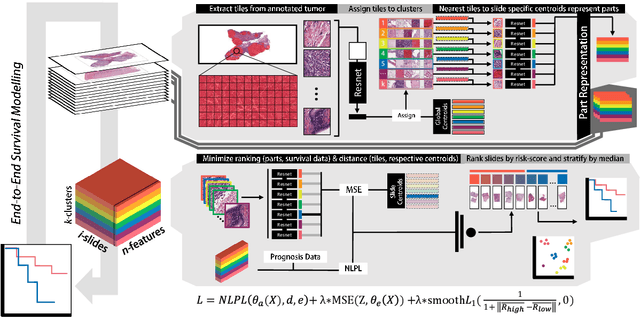
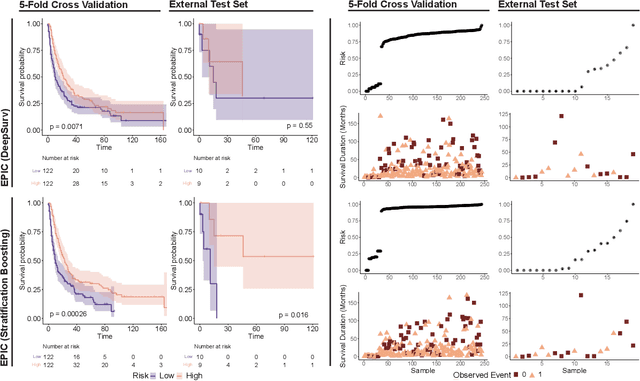
Histopathology-based survival modelling has two major hurdles. Firstly, a well-performing survival model has minimal clinical application if it does not contribute to the stratification of a cancer patient cohort into different risk groups, preferably driven by histologic morphologies. In the clinical setting, individuals are not given specific prognostic predictions, but are rather predicted to lie within a risk group which has a general survival trend. Thus, It is imperative that a survival model produces well-stratified risk groups. Secondly, until now, survival modelling was done in a two-stage approach (encoding and aggregation). The massive amount of pixels in digitized whole slide images were never utilized to their fullest extent due to technological constraints on data processing, forcing decoupled learning. EPIC-Survival bridges encoding and aggregation into an end-to-end survival modelling approach, while introducing stratification boosting to encourage the model to not only optimize ranking, but also to discriminate between risk groups. In this study we show that EPIC-Survival performs better than other approaches in modelling intrahepatic cholangiocarcinoma, a historically difficult cancer to model. Further, we show that stratification boosting improves further improves model performance, resulting in a concordance-index of 0.880 on a held-out test set. Finally, we were able to identify specific histologic differences, not commonly sought out in ICC, between low and high risk groups.
Deep Interactive Learning: An Efficient Labeling Approach for Deep Learning-Based Osteosarcoma Treatment Response Assessment
Jul 02, 2020David Joon Ho, Narasimhan P. Agaram, Peter J. Schueffler, Chad M. Vanderbilt, Marc-Henri Jean, Meera R. Hameed, Thomas J. Fuchs
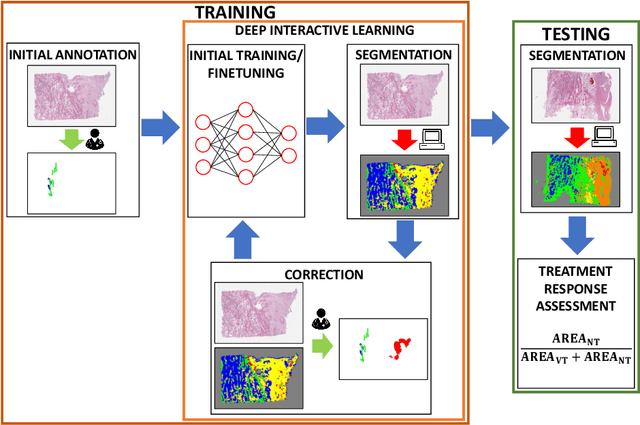
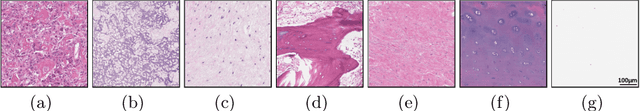
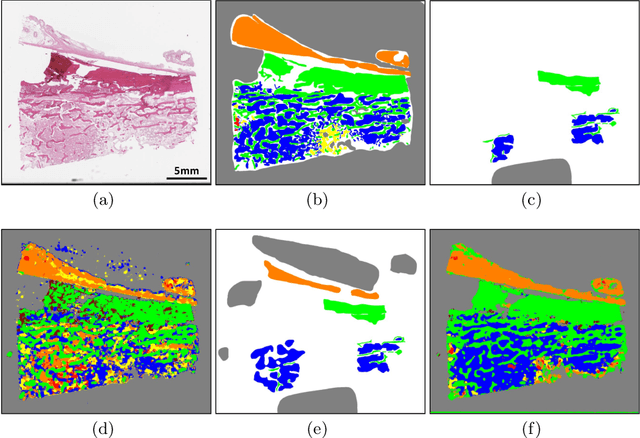

Osteosarcoma is the most common malignant primary bone tumor. Standard treatment includes pre-operative chemotherapy followed by surgical resection. The response to treatment as measured by ratio of necrotic tumor area to overall tumor area is a known prognostic factor for overall survival. This assessment is currently done manually by pathologists by looking at glass slides under the microscope which may not be reproducible due to its subjective nature. Convolutional neural networks (CNNs) can be used for automated segmentation of viable and necrotic tumor on osteosarcoma whole slide images. One bottleneck for supervised learning is that large amounts of accurate annotations are required for training which is a time-consuming and expensive process. In this paper, we describe Deep Interactive Learning (DIaL) as an efficient labeling approach for training CNNs. After an initial labeling step is done, annotators only need to correct mislabeled regions from previous segmentation predictions to improve the CNN model until the satisfactory predictions are achieved. Our experiments show that our CNN model trained by only 7 hours of annotation using DIaL can successfully estimate ratios of necrosis within expected inter-observer variation rate for non-standardized manual surgical pathology task.
Deep Multi-Magnification Networks for Multi-Class Breast Cancer Image Segmentation
Oct 29, 2019David Joon Ho, Dig V. K. Yarlagadda, Timothy M. D'Alfonso, Matthew G. Hanna, Anne Grabenstetter, Peter Ntiamoah, Edi Brogi, Lee K. Tan, Thomas J. Fuchs
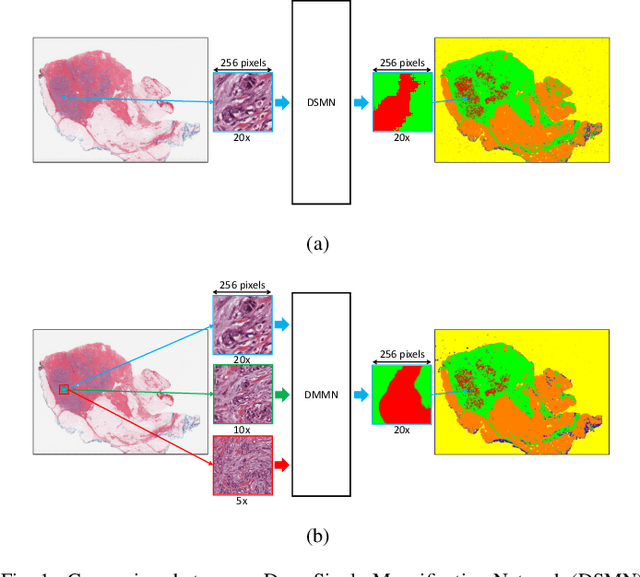
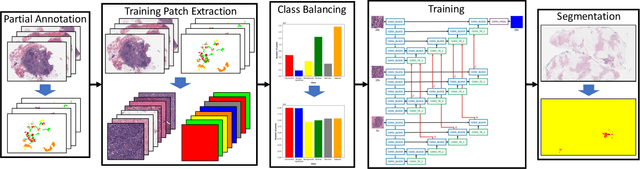
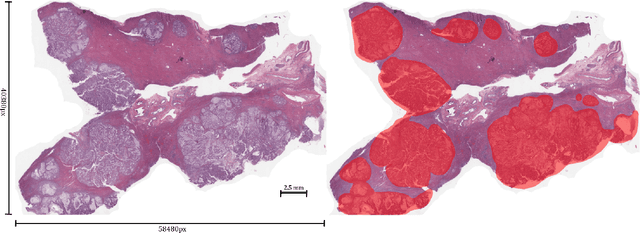
Breast carcinoma is one of the most common cancers for women in the United States. Pathologic analysis of surgical excision specimens for breast carcinoma is important to evaluate the completeness of surgical excision and has implications for future treatment. This analysis is performed manually by pathologists reviewing histologic slides prepared from formalin-fixed tissue. Digital pathology has provided means to digitize the glass slides and generate whole slide images. Computational pathology enables whole slide images to be automatically analyzed to assist pathologists, especially with the advancement of deep learning. The whole slide images generally contain giga-pixels of data, so it is impractical to process the images at the whole-slide-level. Most of the current deep learning techniques process the images at the patch-level, but they may produce poor results by looking at individual patches with a narrow field-of-view at a single magnification. In this paper, we present Deep Multi-Magnification Networks (DMMNs) to resemble how pathologists analyze histologic slides using microscopes. Our multi-class tissue segmentation architecture processes a set of patches from multiple magnifications to make more accurate predictions. For our supervised training, we use partial annotations to reduce the burden of annotators. Our segmentation architecture with multi-encoder, multi-decoder, and multi-concatenation outperforms other segmentation architectures on breast datasets and can be used to facilitate pathologists' assessments of breast cancer.
Towards Unsupervised Cancer Subtyping: Predicting Prognosis Using A Histologic Visual Dictionary
Mar 12, 2019Hassan Muhammad, Carlie S. Sigel, Gabriele Campanella, Thomas Boerner, Linda M. Pak, Stefan Büttner, Jan N. M. IJzermans, Bas Groot Koerkamp, Michael Doukas, William R. Jarnagin, Amber Simpson, Thomas J. Fuchs
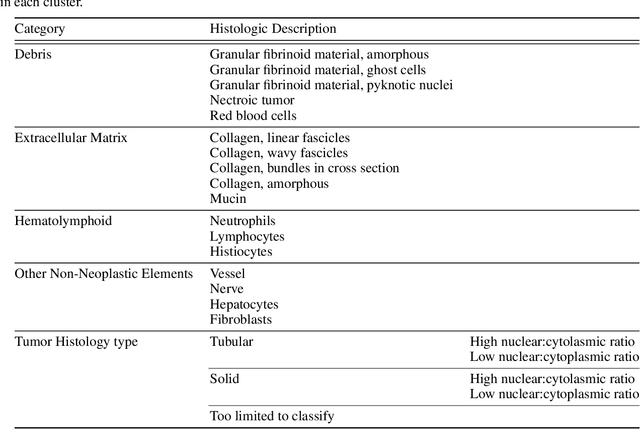
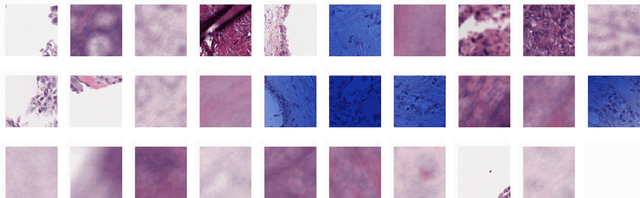
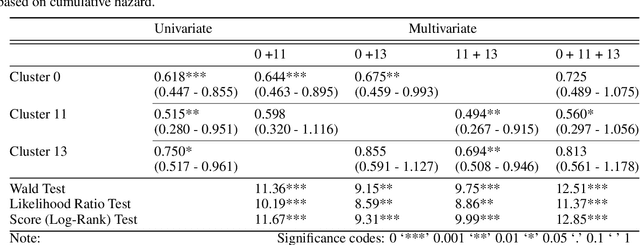
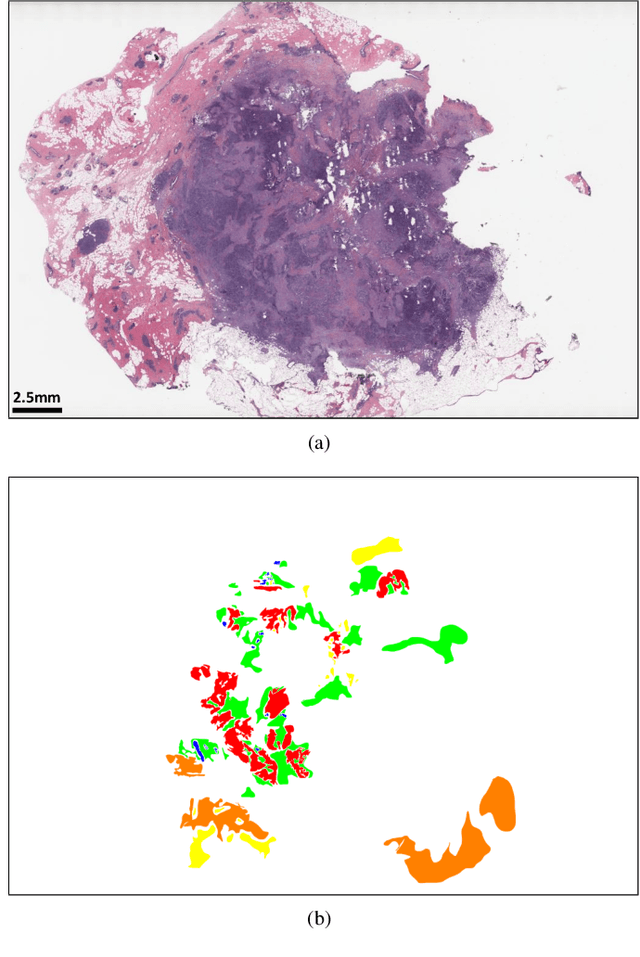
Unlike common cancers, such as those of the prostate and breast, tumor grading in rare cancers is difficult and largely undefined because of small sample sizes, the sheer volume of time needed to undertake on such a task, and the inherent difficulty of extracting human-observed patterns. One of the most challenging examples is intrahepatic cholangiocarcinoma (ICC), a primary liver cancer arising from the biliary system, for which there is well-recognized tumor heterogeneity and no grading paradigm or prognostic biomarkers. In this paper, we propose a new unsupervised deep convolutional autoencoder-based clustering model that groups together cellular and structural morphologies of tumor in 246 ICC digitized whole slides, based on visual similarity. From this visual dictionary of histologic patterns, we use the clusters as covariates to train Cox-proportional hazard survival models. In univariate analysis, three clusters were significantly associated with recurrence-free survival. Combinations of these clusters were significant in multivariate analysis. In a multivariate analysis of all clusters, five showed significance to recurrence-free survival, however the overall model was not measured to be significant. Finally, a pathologist assigned clinical terminology to the significant clusters in the visual dictionary and found evidence supporting the hypothesis that collagen-enriched fibrosis plays a role in disease severity. These results offer insight into the future of cancer subtyping and show that computational pathology can contribute to disease prognostication, especially in rare cancers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge