CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023Simon Graham, Quoc Dang Vu, Mostafa Jahanifar, Martin Weigert, Uwe Schmidt, Wenhua Zhang, Jun Zhang, Sen Yang, Jinxi Xiang, Xiyue Wang, Josef Lorenz Rumberger, Elias Baumann, Peter Hirsch, Lihao Liu, Chenyang Hong, Angelica I. Aviles-Rivero, Ayushi Jain, Heeyoung Ahn, Yiyu Hong, Hussam Azzuni, Min Xu, Mohammad Yaqub, Marie-Claire Blache, Benoît Piégu, Bertrand Vernay, Tim Scherr, Moritz Böhland, Katharina Löffler, Jiachen Li, Weiqin Ying, Chixin Wang, Dagmar Kainmueller, Carola-Bibiane Schönlieb, Shuolin Liu, Dhairya Talsania, Yughender Meda, Prakash Mishra, Muhammad Ridzuan, Oliver Neumann, Marcel P. Schilling, Markus Reischl, Ralf Mikut, Banban Huang, Hsiang-Chin Chien, Ching-Ping Wang, Chia-Yen Lee, Hong-Kun Lin, Zaiyi Liu, Xipeng Pan, Chu Han, Jijun Cheng, Muhammad Dawood, Srijay Deshpande, Raja Muhammad Saad Bashir, Adam Shephard, Pedro Costa, João D. Nunes, Aurélio Campilho, Jaime S. Cardoso, Hrishikesh P S, Densen Puthussery, Devika R G, Jiji C V, Ye Zhang, Zijie Fang, Zhifan Lin, Yongbing Zhang, Chunhui Lin, Liukun Zhang, Lijian Mao, Min Wu, Vi Thi-Tuong Vo, Soo-Hyung Kim, Taebum Lee, Satoshi Kondo, Satoshi Kasai, Pranay Dumbhare, Vedant Phuse, Yash Dubey, Ankush Jamthikar, Trinh Thi Le Vuong, Jin Tae Kwak, Dorsa Ziaei, Hyun Jung, Tianyi Miao, David Snead, Shan E Ahmed Raza, Fayyaz Minhas, Nasir M. Rajpoot
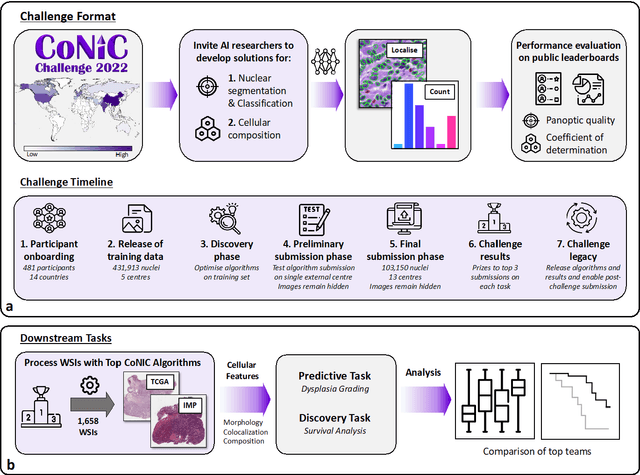
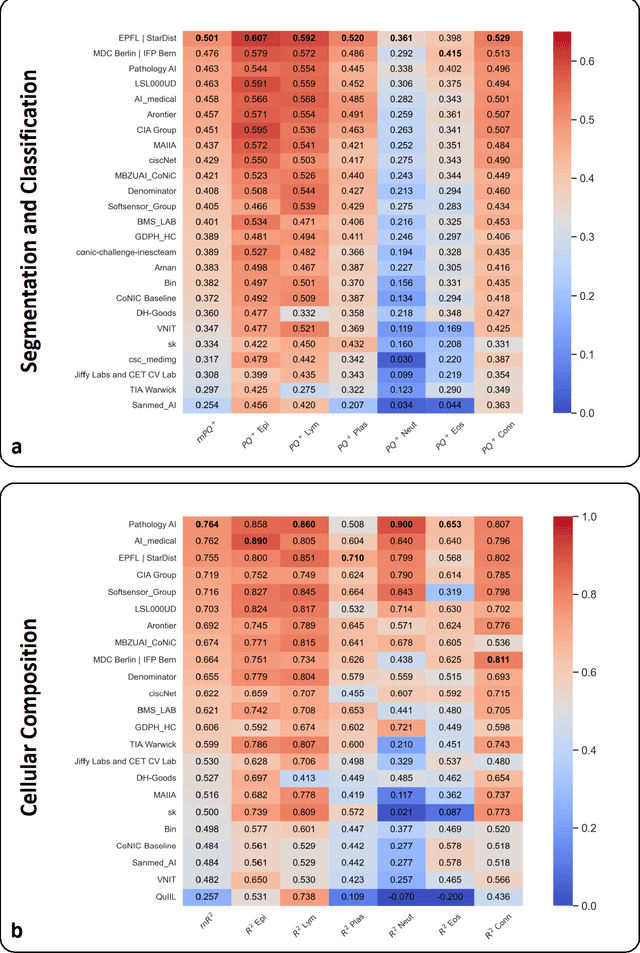
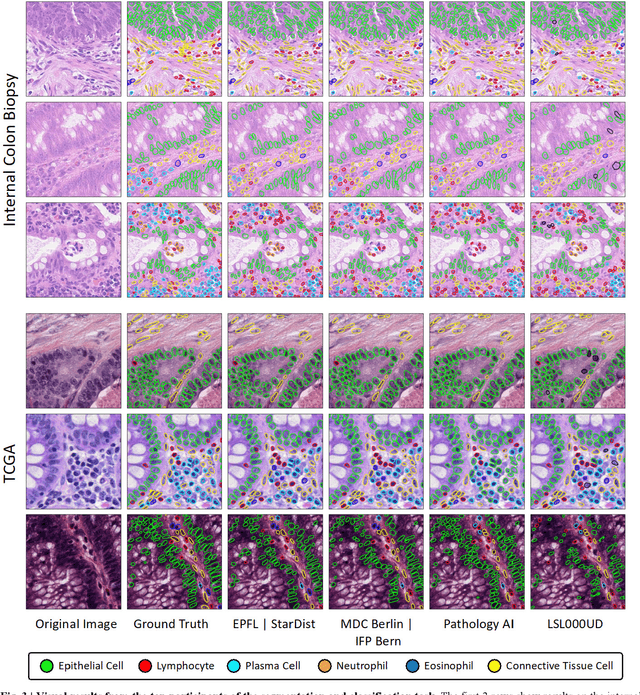
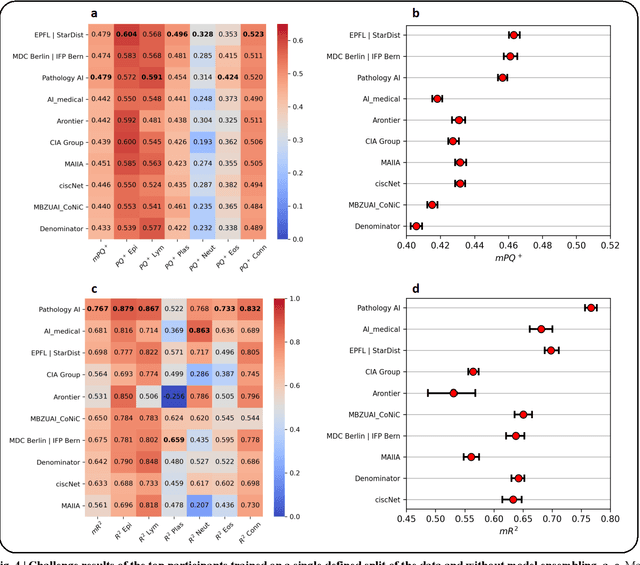
Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Mimicking a Pathologist: Dual Attention Model for Scoring of Gigapixel Histology Images
Feb 19, 2023Manahil Raza, Ruqayya Awan, Raja Muhammad Saad Bashir, Talha Qaiser, Nasir M. Rajpoot
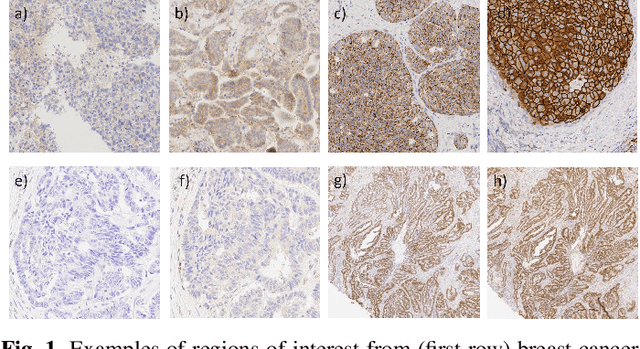
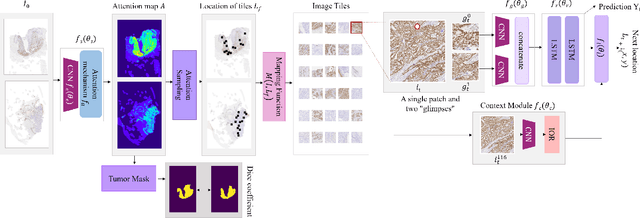
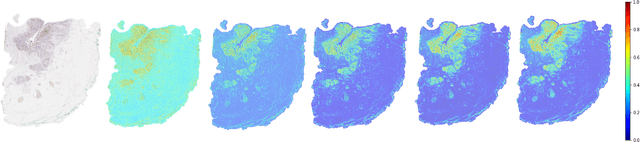
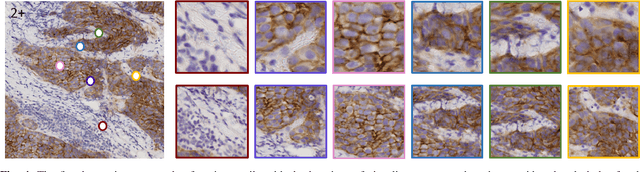
Some major challenges associated with the automated processing of whole slide images (WSIs) includes their sheer size, different magnification levels and high resolution. Utilizing these images directly in AI frameworks is computationally expensive due to memory constraints, while downsampling WSIs incurs information loss and splitting WSIs into tiles and patches results in loss of important contextual information. We propose a novel dual attention approach, consisting of two main components, to mimic visual examination by a pathologist. The first component is a soft attention model which takes as input a high-level view of the WSI to determine various regions of interest. We employ a custom sampling method to extract diverse and spatially distinct image tiles from selected high attention areas. The second component is a hard attention classification model, which further extracts a sequence of multi-resolution glimpses from each tile for classification. Since hard attention is non-differentiable, we train this component using reinforcement learning and predict the location of glimpses without processing all patches of a given tile, thereby aligning with pathologist's way of diagnosis. We train our components both separately and in an end-to-end fashion using a joint loss function to demonstrate the efficacy of our proposed model. We employ our proposed model on two different IHC use cases: HER2 prediction on breast cancer and prediction of Intact/Loss status of two MMR biomarkers, for colorectal cancer. We show that the proposed model achieves accuracy comparable to state-of-the-art methods while only processing a small fraction of the WSI at highest magnification.
Consistency Regularisation in Varying Contexts and Feature Perturbations for Semi-Supervised Semantic Segmentation of Histology Images
Feb 11, 2023Raja Muhammad Saad Bashir, Talha Qaiser, Shan E Ahmed Raza, Nasir M. Rajpoot
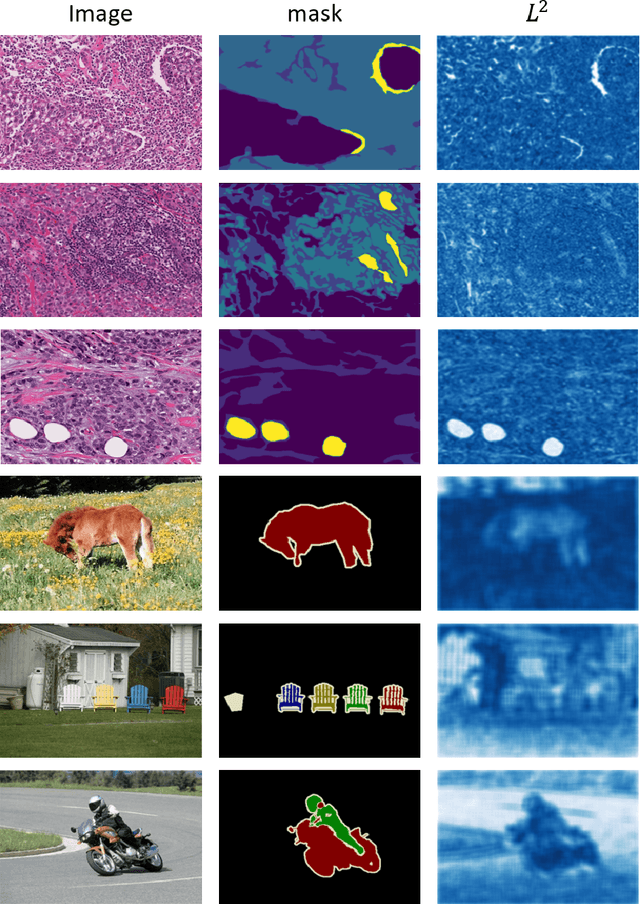
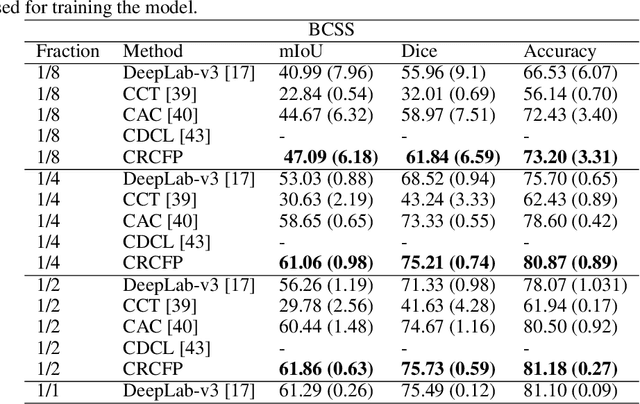
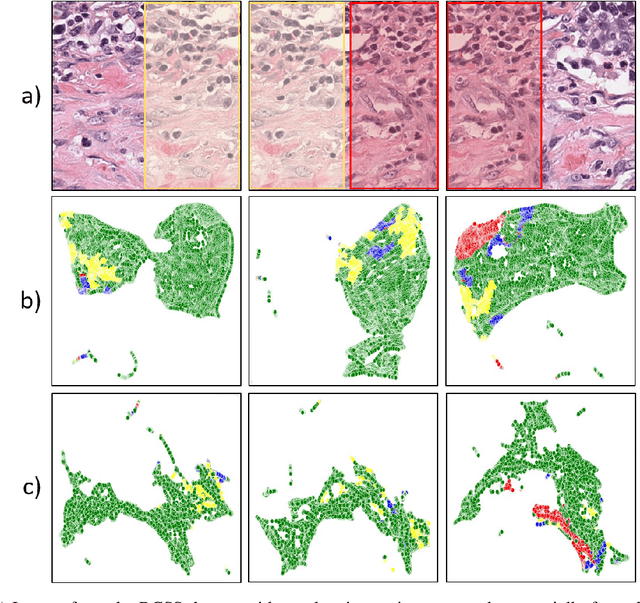
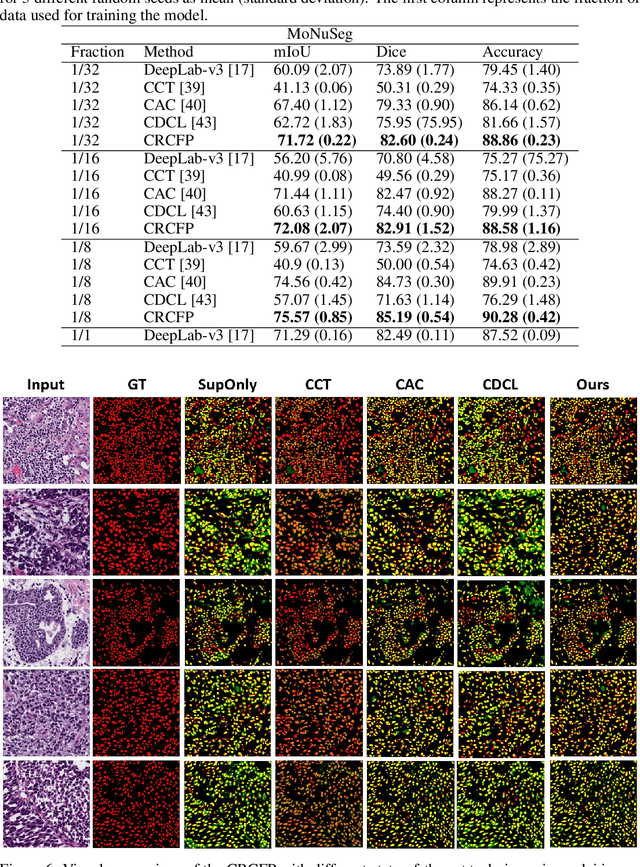
Semantic segmentation of various tissue and nuclei types in histology images is fundamental to many downstream tasks in the area of computational pathology (CPath). In recent years, Deep Learning (DL) methods have been shown to perform well on segmentation tasks but DL methods generally require a large amount of pixel-wise annotated data. Pixel-wise annotation sometimes requires expert's knowledge and time which is laborious and costly to obtain. In this paper, we present a consistency based semi-supervised learning (SSL) approach that can help mitigate this challenge by exploiting a large amount of unlabelled data for model training thus alleviating the need for a large annotated dataset. However, SSL models might also be susceptible to changing context and features perturbations exhibiting poor generalisation due to the limited training data. We propose an SSL method that learns robust features from both labelled and unlabelled images by enforcing consistency against varying contexts and feature perturbations. The proposed method incorporates context-aware consistency by contrasting pairs of overlapping images in a pixel-wise manner from changing contexts resulting in robust and context invariant features. We show that cross-consistency training makes the encoder features invariant to different perturbations and improves the prediction confidence. Finally, entropy minimisation is employed to further boost the confidence of the final prediction maps from unlabelled data. We conduct an extensive set of experiments on two publicly available large datasets (BCSS and MoNuSeg) and show superior performance compared to the state-of-the-art methods.
Deep Feature based Cross-slide Registration
Feb 27, 2022Ruqayya Awan, Shan E Ahmed Raza, Johannes Lotz, Nick Weiss, Nasir M. Rajpoot
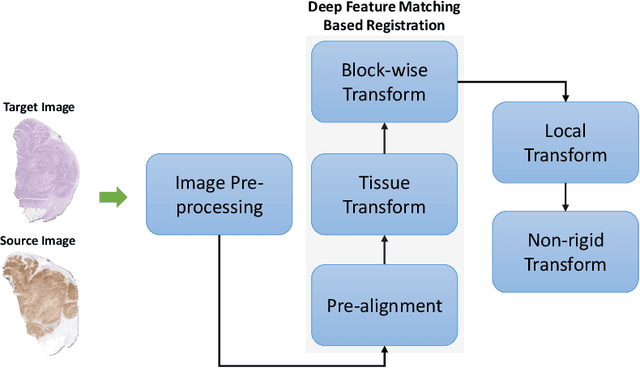
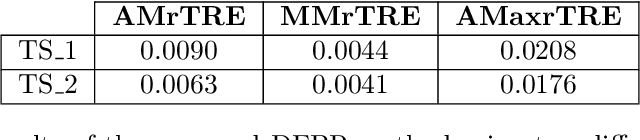
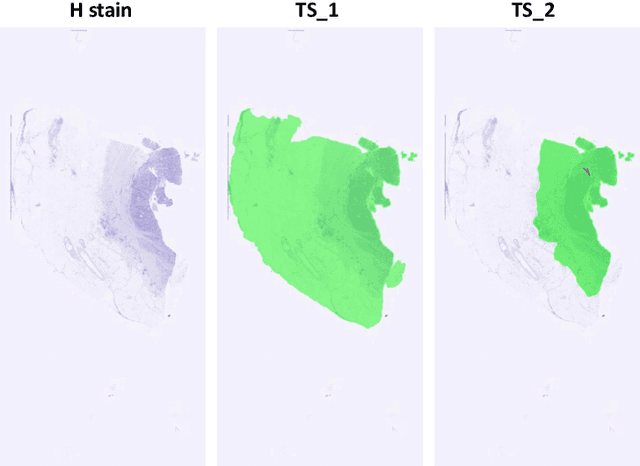
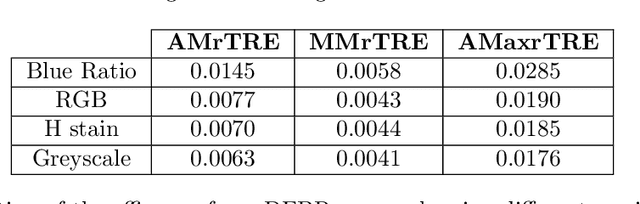
Cross-slide image analysis provides additional information by analysing the expression of different biomarkers as compared to a single slide analysis. Slides stained with different biomarkers are analysed side by side which may reveal unknown relations between the different biomarkers. During the slide preparation, a tissue section may be placed at an arbitrary orientation as compared to other sections of the same tissue block. The problem is compounded by the fact that tissue contents are likely to change from one section to the next and there may be unique artefacts on some of the slides. This makes registration of each section to a reference section of the same tissue block an important pre-requisite task before any cross-slide analysis. We propose a deep feature based registration (DFBR) method which utilises data-driven features to estimate the rigid transformation. We adopted a multi-stage strategy for improving the quality of registration. We also developed a visualisation tool to view registered pairs of WSIs at different magnifications. With the help of this tool, one can apply a transformation on the fly without the need to generate transformed source WSI in a pyramidal form. We compared the performance of data-driven features with that of hand-crafted features on the COMET dataset. Our approach can align the images with low registration errors. Generally, the success of non-rigid registration is dependent on the quality of rigid registration. To evaluate the efficacy of the DFBR method, the first two steps of the ANHIR winner's framework are replaced with our DFBR to register challenge provided image pairs. The modified framework produce comparable results to that of challenge winning team.
Deep Learning based Prediction of MSI in Colorectal Cancer via Prediction of the Status of MMR Markers
Feb 24, 2022Ruqayya Awan, Mohammed Nimir, Shan E Ahmed Raza, Johannes Lotz, David Snead, Andrew Robison, Nasir M. Rajpoot
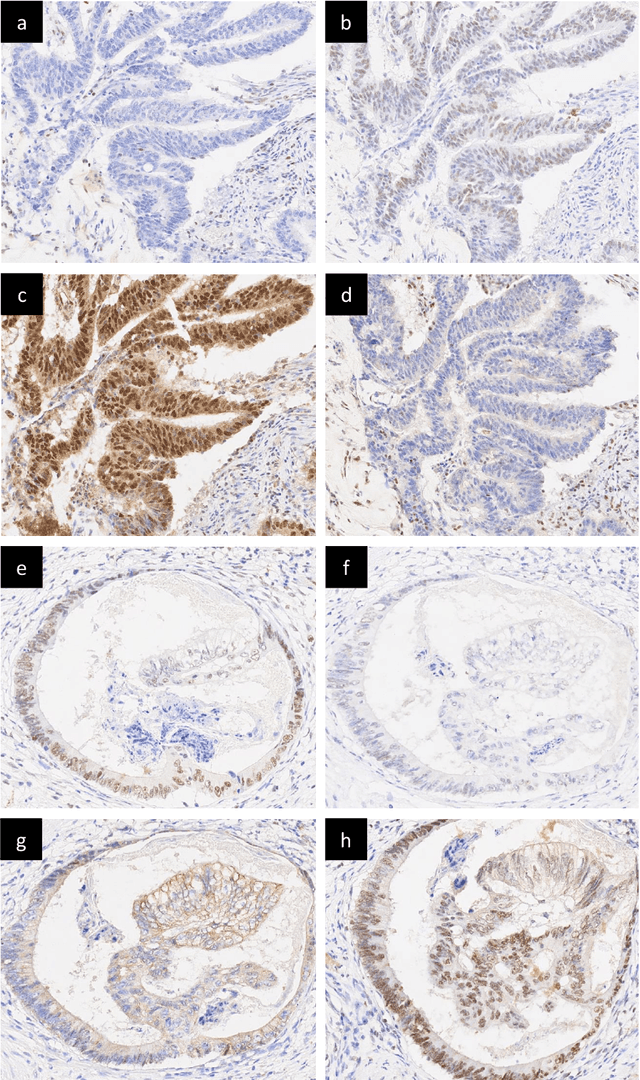

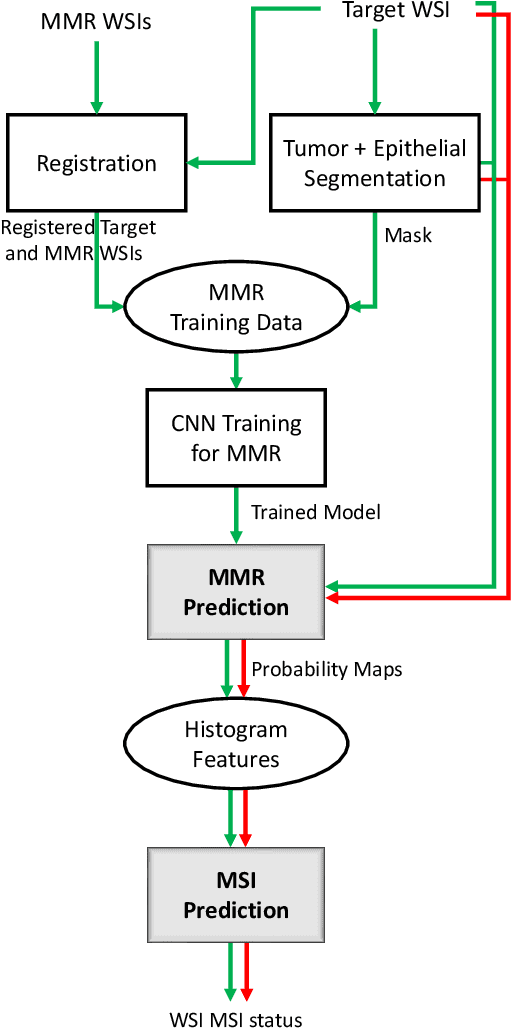
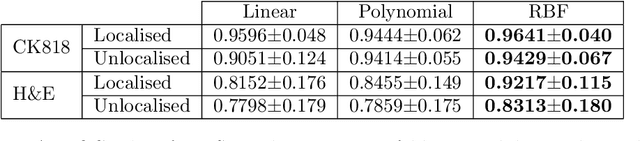
An accurate diagnosis and profiling of tumour are critical to the best treatment choices for cancer patients. In addition to the cancer type and its aggressiveness, molecular heterogeneity also plays a vital role in treatment selection. MSI or MMR deficiency is one of the well-studied aberrations in terms of molecular changes. Colorectal cancer patients with MMR deficiency respond well to immunotherapy, hence assessment of the relevant molecular markers can assist clinicians in making optimal treatment selections for patients. Immunohistochemistry is one of the ways for identifying these molecular changes which requires additional sections of tumour tissue. Introduction of automated methods that can predict MSI or MMR status from a target image without the need for additional sections can substantially reduce the cost associated with it. In this work, we present our work on predicting MSI status in a two-stage process using a single target slide either stained with CK818 or H\&E. First, we train a multi-headed convolutional neural network model where each head is responsible for predicting one of the MMR protein expressions. To this end, we perform registration of MMR slides to the target slide as a pre-processing step. In the second stage, statistical features computed from the MMR prediction maps are used for the final MSI prediction. Our results demonstrate that MSI classification can be improved on incorporating fine-grained MMR labels in comparison to the previous approaches in which coarse labels (MSI/MSS) are utilised.
FedDropoutAvg: Generalizable federated learning for histopathology image classification
Nov 25, 2021Gozde N. Gunesli, Mohsin Bilal, Shan E Ahmed Raza, Nasir M. Rajpoot
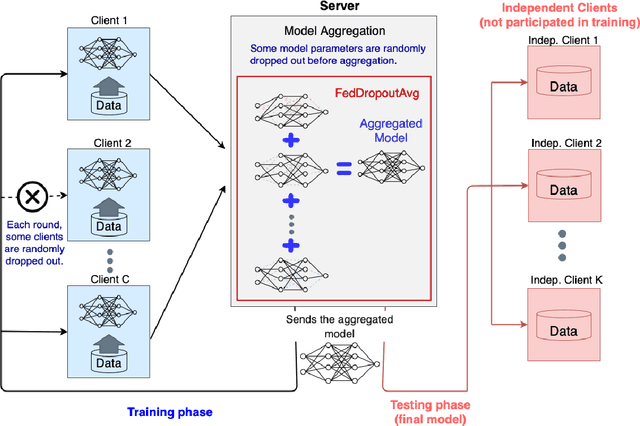
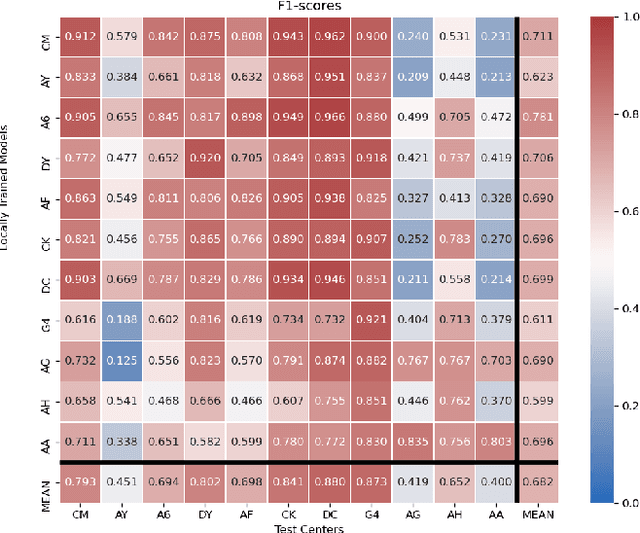
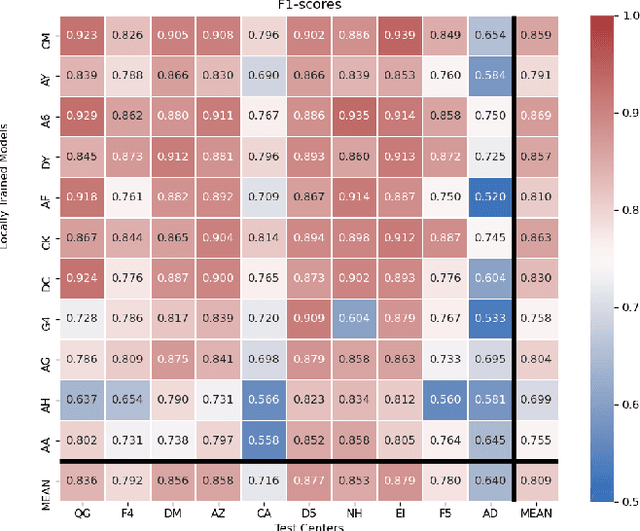
Federated learning (FL) enables collaborative learning of a deep learning model without sharing the data of participating sites. FL in medical image analysis tasks is relatively new and open for enhancements. In this study, we propose FedDropoutAvg, a new federated learning approach for training a generalizable model. The proposed method takes advantage of randomness, both in client selection and also in federated averaging process. We compare FedDropoutAvg to several algorithms in an FL scenario for real-world multi-site histopathology image classification task. We show that with FedDropoutAvg, the final model can achieve performance better than other FL approaches and closer to a classical deep learning model that requires all data to be shared for centralized training. We test the trained models on a large dataset consisting of 1.2 million image tiles from 21 different centers. To evaluate the generalization ability of the proposed approach, we use held-out test sets from centers whose data was used in the FL and for unseen data from other independent centers whose data was not used in the federated training. We show that the proposed approach is more generalizable than other state-of-the-art federated training approaches. To the best of our knowledge, ours is the first study to use a randomized client and local model parameter selection procedure in a federated setting for a medical image analysis task.
Simultaneous Nuclear Instance and Layer Segmentation in Oral Epithelial Dysplasia
Sep 01, 2021Adam J. Shephard, Simon Graham, R. M. Saad Bashir, Mostafa Jahanifar, Hanya Mahmood, Syed Ali Khurram, Nasir M. Rajpoot
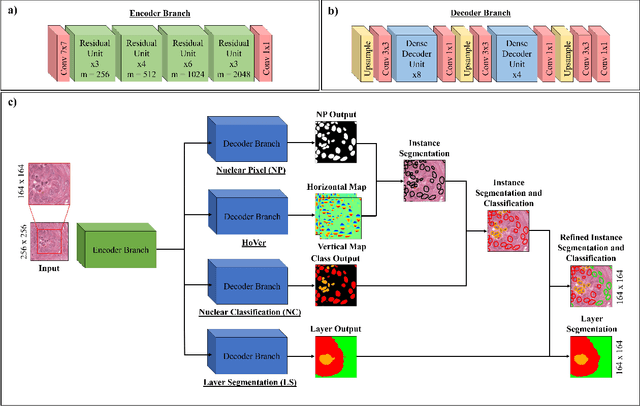
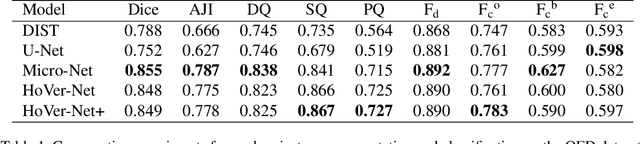
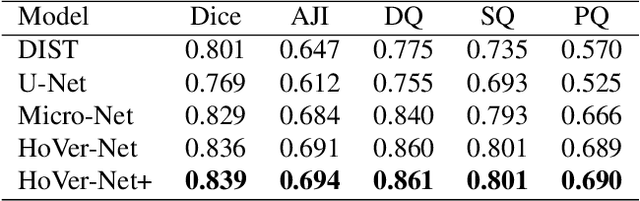
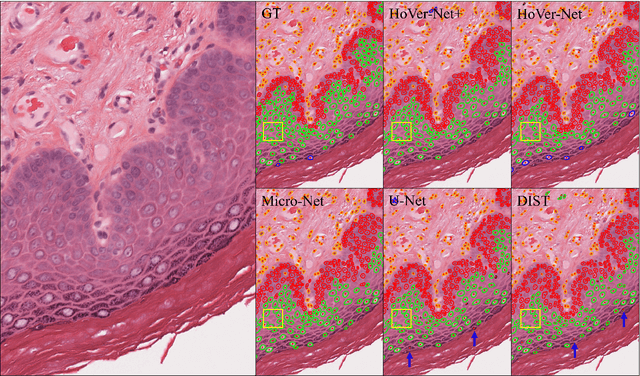
Oral epithelial dysplasia (OED) is a pre-malignant histopathological diagnosis given to lesions of the oral cavity. Predicting OED grade or whether a case will transition to malignancy is critical for early detection and appropriate treatment. OED typically begins in the lower third of the epithelium before progressing upwards with grade severity, thus we have suggested that segmenting intra-epithelial layers, in addition to individual nuclei, may enable researchers to evaluate important layer-specific morphological features for grade/malignancy prediction. We present HoVer-Net+, a deep learning framework to simultaneously segment (and classify) nuclei and (intra-)epithelial layers in H&E stained slides from OED cases. The proposed architecture consists of an encoder branch and four decoder branches for simultaneous instance segmentation of nuclei and semantic segmentation of the epithelial layers. We show that the proposed model achieves the state-of-the-art (SOTA) performance in both tasks, with no additional costs when compared to previous SOTA methods for each task. To the best of our knowledge, ours is the first method for simultaneous nuclear instance segmentation and semantic tissue segmentation, with potential for use in computational pathology for other similar simultaneous tasks and for future studies into malignancy prediction.
ALBRT: Cellular Composition Prediction in Routine Histology Images
Aug 26, 2021Muhammad Dawood, Kim Branson, Nasir M. Rajpoot, Fayyaz ul Amir Afsar Minhas
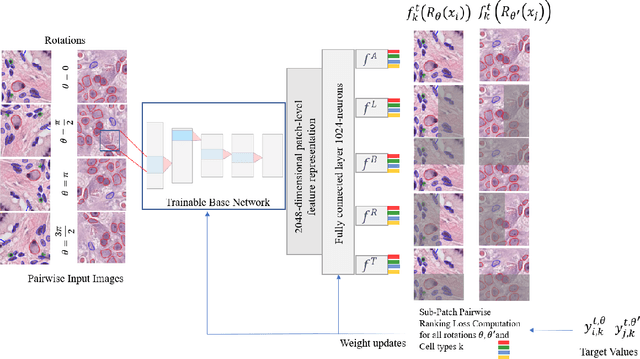
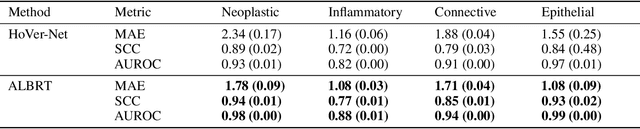
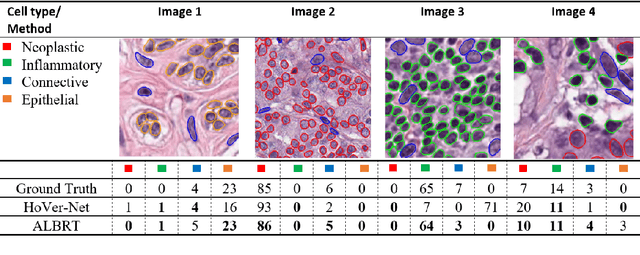
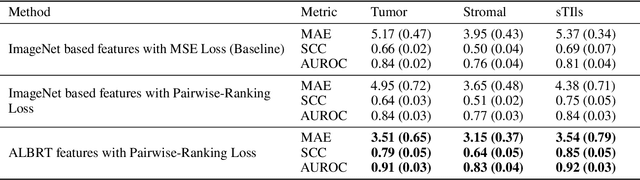
Cellular composition prediction, i.e., predicting the presence and counts of different types of cells in the tumor microenvironment from a digitized image of a Hematoxylin and Eosin (H&E) stained tissue section can be used for various tasks in computational pathology such as the analysis of cellular topology and interactions, subtype prediction, survival analysis, etc. In this work, we propose an image-based cellular composition predictor (ALBRT) which can accurately predict the presence and counts of different types of cells in a given image patch. ALBRT, by its contrastive-learning inspired design, learns a compact and rotation-invariant feature representation that is then used for cellular composition prediction of different cell types. It offers significant improvement over existing state-of-the-art approaches for cell classification and counting. The patch-level feature representation learned by ALBRT is transferrable for cellular composition analysis over novel datasets and can also be utilized for downstream prediction tasks in CPath as well. The code and the inference webserver for the proposed method are available at the URL: https://github.com/engrodawood/ALBRT.
All You Need is Color: Image based Spatial Gene Expression Prediction using Neural Stain Learning
Aug 26, 2021Muhammad Dawood, Kim Branson, Nasir M. Rajpoot, Fayyaz ul Amir Afsar Minhas
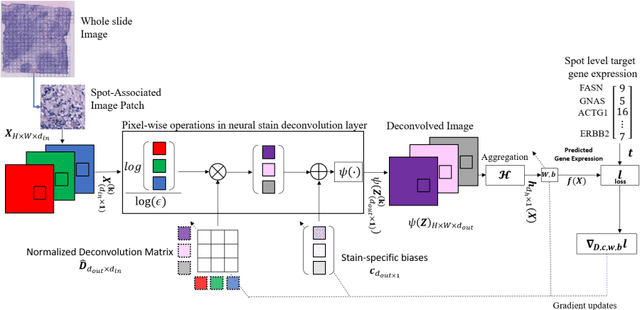
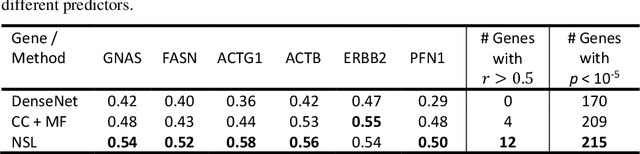
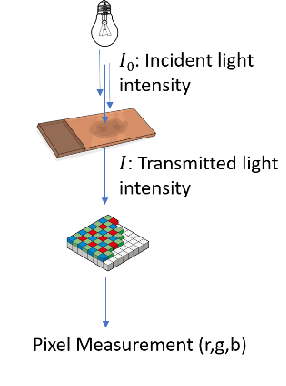
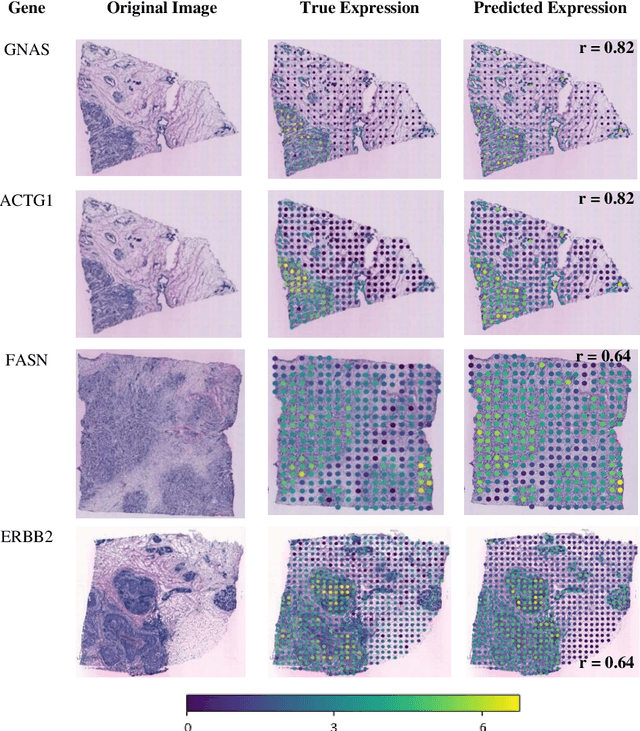
"Is it possible to predict expression levels of different genes at a given spatial location in the routine histology image of a tumor section by modeling its stain absorption characteristics?" In this work, we propose a "stain-aware" machine learning approach for prediction of spatial transcriptomic gene expression profiles using digital pathology image of a routine Hematoxylin & Eosin (H&E) histology section. Unlike recent deep learning methods which are used for gene expression prediction, our proposed approach termed Neural Stain Learning (NSL) explicitly models the association of stain absorption characteristics of the tissue with gene expression patterns in spatial transcriptomics by learning a problem-specific stain deconvolution matrix in an end-to-end manner. The proposed method with only 11 trainable weight parameters outperforms both classical regression models with cellular composition and morphological features as well as deep learning methods. We have found that the gene expression predictions from the proposed approach show higher correlations with true expression values obtained through sequencing for a larger set of genes in comparison to other approaches.
L1-regularized neural ranking for risk stratification and its application to prediction of time to distant metastasis in luminal node negative chemotherapy naïve breast cancer patients
Aug 23, 2021Fayyaz Minhas, Michael S. Toss, Noor ul Wahab, Emad Rakha, Nasir M. Rajpoot
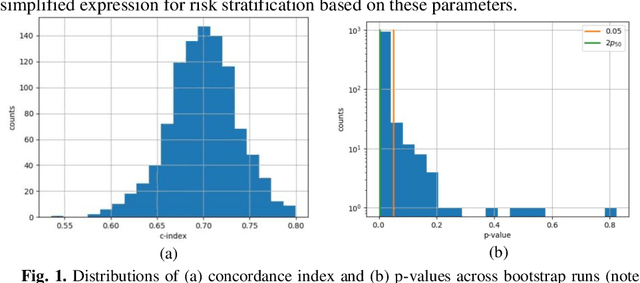

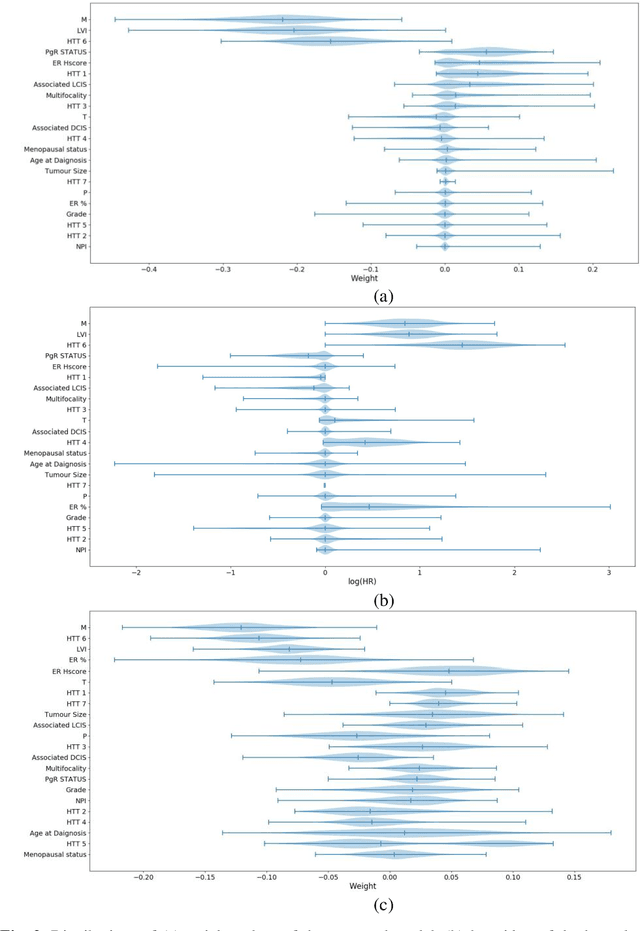
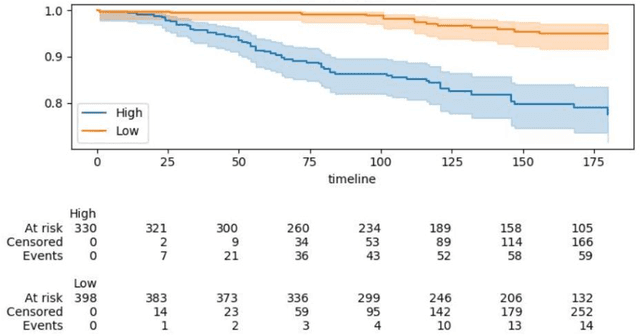
Can we predict if an early stage cancer patient is at high risk of developing distant metastasis and what clinicopathological factors are associated with such a risk? In this paper, we propose a ranking based censoring-aware machine learning model for answering such questions. The proposed model is able to generate an interpretable formula for risk stratifi-cation using a minimal number of clinicopathological covariates through L1-regulrization. Using this approach, we analyze the association of time to distant metastasis (TTDM) with various clinical parameters for early stage, luminal (ER+ or HER2-) breast cancer patients who received endocrine therapy but no chemotherapy (n = 728). The TTDM risk stratification formula obtained using the proposed approach is primarily based on mitotic score, histolog-ical tumor type and lymphovascular invasion. These findings corroborate with the known role of these covariates in increased risk for distant metastasis. Our analysis shows that the proposed risk stratification formula can discriminate between cases with high and low risk of distant metastasis (p-value < 0.005) and can also rank cases based on their time to distant metastasis with a concordance-index of 0.73.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge