MM-SurvNet: Deep Learning-Based Survival Risk Stratification in Breast Cancer Through Multimodal Data Fusion
Feb 19, 2024Raktim Kumar Mondol, Ewan K. A. Millar, Arcot Sowmya, Erik Meijering
Survival risk stratification is an important step in clinical decision making for breast cancer management. We propose a novel deep learning approach for this purpose by integrating histopathological imaging, genetic and clinical data. It employs vision transformers, specifically the MaxViT model, for image feature extraction, and self-attention to capture intricate image relationships at the patient level. A dual cross-attention mechanism fuses these features with genetic data, while clinical data is incorporated at the final layer to enhance predictive accuracy. Experiments on the public TCGA-BRCA dataset show that our model, trained using the negative log likelihood loss function, can achieve superior performance with a mean C-index of 0.64, surpassing existing methods. This advancement facilitates tailored treatment strategies, potentially leading to improved patient outcomes.
BioFusionNet: Deep Learning-Based Survival Risk Stratification in ER+ Breast Cancer Through Multifeature and Multimodal Data Fusion
Feb 16, 2024Raktim Kumar Mondol, Ewan K. A. Millar, Arcot Sowmya, Erik Meijering
Breast cancer is a significant health concern affecting millions of women worldwide. Accurate survival risk stratification plays a crucial role in guiding personalised treatment decisions and improving patient outcomes. Here we present BioFusionNet, a deep learning framework that fuses image-derived features with genetic and clinical data to achieve a holistic patient profile and perform survival risk stratification of ER+ breast cancer patients. We employ multiple self-supervised feature extractors, namely DINO and MoCoV3, pretrained on histopathology patches to capture detailed histopathological image features. We then utilise a variational autoencoder (VAE) to fuse these features, and harness the latent space of the VAE to feed into a self-attention network, generating patient-level features. Next, we develop a co-dual-cross-attention mechanism to combine the histopathological features with genetic data, enabling the model to capture the interplay between them. Additionally, clinical data is incorporated using a feed-forward network (FFN), further enhancing predictive performance and achieving comprehensive multimodal feature integration. Furthermore, we introduce a weighted Cox loss function, specifically designed to handle imbalanced survival data, which is a common challenge in the field. The proposed model achieves a mean concordance index (C-index) of 0.77 and a time-dependent area under the curve (AUC) of 0.84, outperforming state-of-the-art methods. It predicts risk (high versus low) with prognostic significance for overall survival (OS) in univariate analysis (HR=2.99, 95% CI: 1.88--4.78, p<0.005), and maintains independent significance in multivariate analysis incorporating standard clinicopathological variables (HR=2.91, 95% CI: 1.80--4.68, p<0.005). The proposed method not only improves model performance but also addresses a critical gap in handling imbalanced data.
ESDMR-Net: A Lightweight Network With Expand-Squeeze and Dual Multiscale Residual Connections for Medical Image Segmentation
Dec 17, 2023Tariq M Khan, Syed S. Naqvi, Erik Meijering
Segmentation is an important task in a wide range of computer vision applications, including medical image analysis. Recent years have seen an increase in the complexity of medical image segmentation approaches based on sophisticated convolutional neural network architectures. This progress has led to incremental enhancements in performance on widely recognised benchmark datasets. However, most of the existing approaches are computationally demanding, which limits their practical applicability. This paper presents an expand-squeeze dual multiscale residual network (ESDMR-Net), which is a fully convolutional network that is particularly well-suited for resource-constrained computing hardware such as mobile devices. ESDMR-Net focuses on extracting multiscale features, enabling the learning of contextual dependencies among semantically distinct features. The ESDMR-Net architecture allows dual-stream information flow within encoder-decoder pairs. The expansion operation (depthwise separable convolution) makes all of the rich features with multiscale information available to the squeeze operation (bottleneck layer), which then extracts the necessary information for the segmentation task. The Expand-Squeeze (ES) block helps the network pay more attention to under-represented classes, which contributes to improved segmentation accuracy. To enhance the flow of information across multiple resolutions or scales, we integrated dual multiscale residual (DMR) blocks into the skip connection. This integration enables the decoder to access features from various levels of abstraction, ultimately resulting in more comprehensive feature representations. We present experiments on seven datasets from five distinct examples of applications. Our model achieved the best results despite having significantly fewer trainable parameters, with a reduction of two or even three orders of magnitude.
Feature Enhancer Segmentation Network (FES-Net) for Vessel Segmentation
Sep 07, 2023Tariq M. Khan, Muhammad Arsalan, Shahzaib Iqbal, Imran Razzak, Erik Meijering
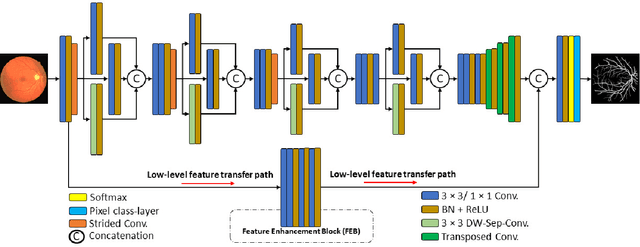
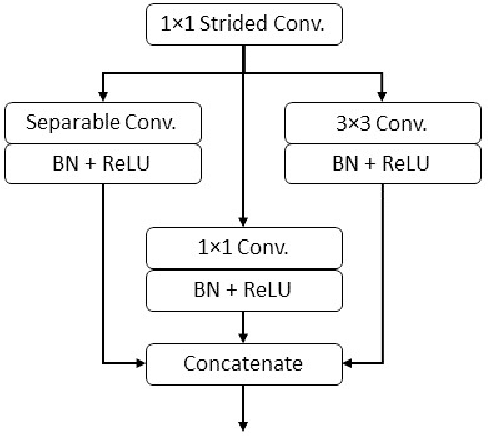
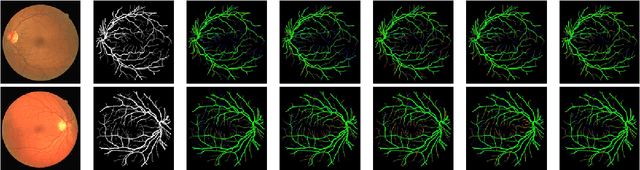
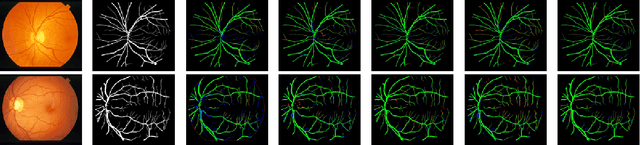
Diseases such as diabetic retinopathy and age-related macular degeneration pose a significant risk to vision, highlighting the importance of precise segmentation of retinal vessels for the tracking and diagnosis of progression. However, existing vessel segmentation methods that heavily rely on encoder-decoder structures struggle to capture contextual information about retinal vessel configurations, leading to challenges in reconciling semantic disparities between encoder and decoder features. To address this, we propose a novel feature enhancement segmentation network (FES-Net) that achieves accurate pixel-wise segmentation without requiring additional image enhancement steps. FES-Net directly processes the input image and utilizes four prompt convolutional blocks (PCBs) during downsampling, complemented by a shallow upsampling approach to generate a binary mask for each class. We evaluate the performance of FES-Net on four publicly available state-of-the-art datasets: DRIVE, STARE, CHASE, and HRF. The evaluation results clearly demonstrate the superior performance of FES-Net compared to other competitive approaches documented in the existing literature.
hist2RNA: An efficient deep learning architecture to predict gene expression from breast cancer histopathology images
May 02, 2023Raktim Kumar Mondol, Ewan K. A. Millar, Peter H Graham, Lois Browne, Arcot Sowmya, Erik Meijering
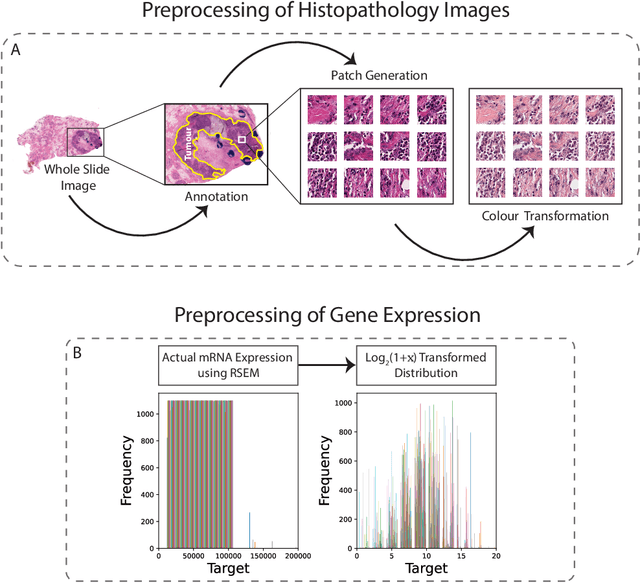
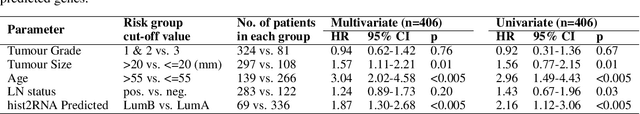
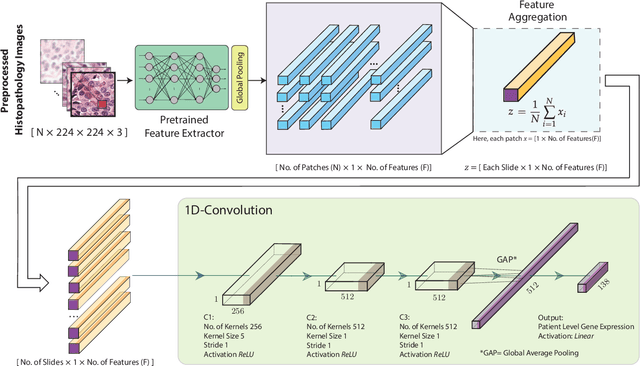
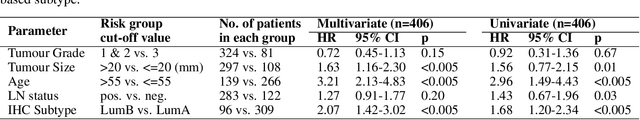
Gene expression can be used to subtype breast cancer with improved prediction of risk of recurrence and treatment responsiveness over that obtained using routine immunohistochemistry (IHC). However, in the clinic, molecular profiling is primarily used for ER+ cancer and is costly and tissue destructive, requires specialized platforms and takes several weeks to obtain a result. Deep learning algorithms can effectively extract morphological patterns in digital histopathology images to predict molecular phenotypes quickly and cost-effectively. We propose a new, computationally efficient approach called hist2RNA inspired by bulk RNA-sequencing techniques to predict the expression of 138 genes (incorporated from six commercially available molecular profiling tests), including luminal PAM50 subtype, from hematoxylin and eosin (H&E) stained whole slide images (WSIs). The training phase involves the aggregation of extracted features for each patient from a pretrained model to predict gene expression at the patient level using annotated H&E images from The Cancer Genome Atlas (TCGA, n=335). We demonstrate successful gene prediction on a held-out test set (n = 160, corr = 0.82 across patients, corr = 0.29 across genes) and perform exploratory analysis on an external tissue microarray (TMA) dataset (n = 498) with known IHC and survival information. Our model is able to predict gene expression and luminal PAM50 subtype (Luminal A versus Luminal B) on the TMA dataset with prognostic significance for overall survival in univariate analysis (c-index = 0.56, hazard ratio = 2.16 (95% CI 1.12-3.06), p < 5 x 10-3), and independent significance in multivariate analysis incorporating standard clinicopathological variables (c-index = 0.65, hazard ratio = 1.85 (95% CI 1.30-2.68), p < 5 x 10-3).
* 16 pages, 10 figures, 2 tables
Breast Cancer Histopathology Image based Gene Expression Prediction using Spatial Transcriptomics data and Deep Learning
Mar 17, 2023Md Mamunur Rahaman, Ewan K. A. Millar, Erik Meijering
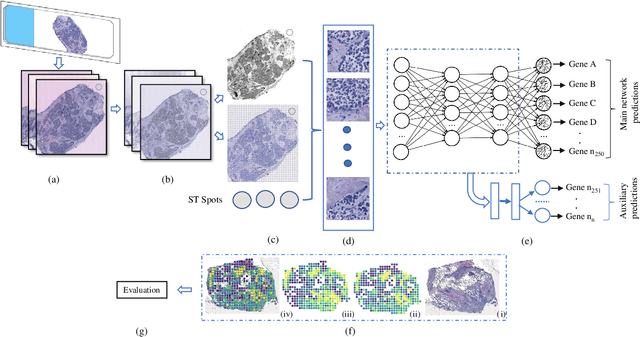

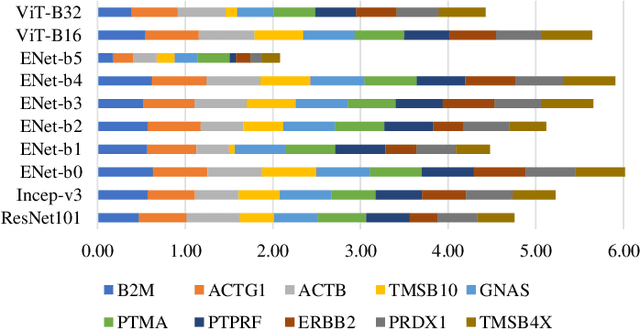
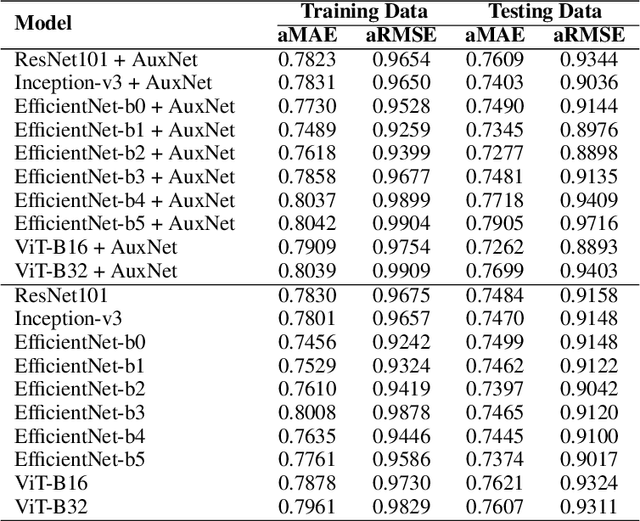
Tumour heterogeneity in breast cancer poses challenges in predicting outcome and response to therapy. Spatial transcriptomics technologies may address these challenges, as they provide a wealth of information about gene expression at the cell level, but they are expensive, hindering their use in large-scale clinical oncology studies. Predicting gene expression from hematoxylin and eosin stained histology images provides a more affordable alternative for such studies. Here we present BrST-Net, a deep learning framework for predicting gene expression from histopathology images using spatial transcriptomics data. Using this framework, we trained and evaluated 10 state-of-the-art deep learning models without utilizing pretrained weights for the prediction of 250 genes. To enhance the generalisation performance of the main network, we introduce an auxiliary network into the framework. Our methodology outperforms previous studies, with 237 genes identified with positive correlation, including 24 genes with a median correlation coefficient greater than 0.50. This is a notable improvement over previous studies, which could predict only 102 genes with positive correlation, with the highest correlation values ranging from 0.29 to 0.34.
Hybrid Dual Mean-Teacher Network With Double-Uncertainty Guidance for Semi-Supervised Segmentation of MRI Scans
Mar 09, 2023Jiayi Zhu, Bart Bolsterlee, Brian V. Y. Chow, Yang Song, Erik Meijering

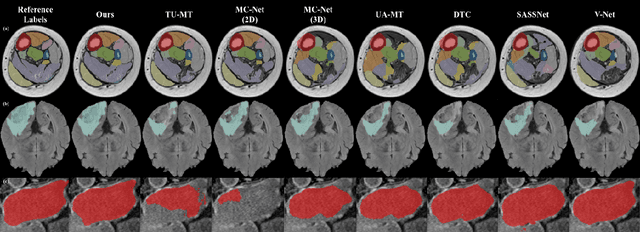
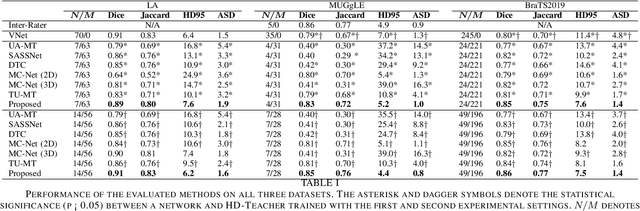
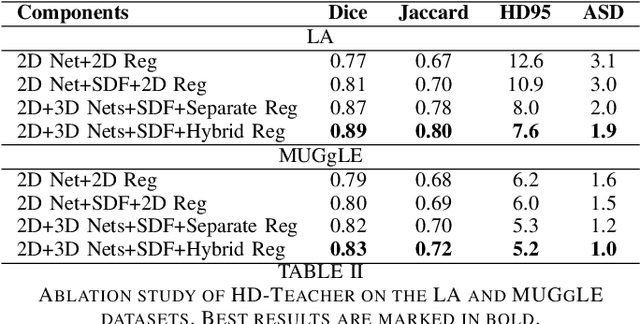
Semi-supervised learning has made significant progress in medical image segmentation. However, existing methods primarily utilize information acquired from a single dimensionality (2D/3D), resulting in sub-optimal performance on challenging data, such as magnetic resonance imaging (MRI) scans with multiple objects and highly anisotropic resolution. To address this issue, we present a Hybrid Dual Mean-Teacher (HD-Teacher) model with hybrid, semi-supervised, and multi-task learning to achieve highly effective semi-supervised segmentation. HD-Teacher employs a 2D and a 3D mean-teacher network to produce segmentation labels and signed distance fields from the hybrid information captured in both dimensionalities. This hybrid learning mechanism allows HD-Teacher to combine the `best of both worlds', utilizing features extracted from either 2D, 3D, or both dimensions to produce outputs as it sees fit. Outputs from 2D and 3D teacher models are also dynamically combined, based on their individual uncertainty scores, into a single hybrid prediction, where the hybrid uncertainty is estimated. We then propose a hybrid regularization module to encourage both student models to produce results close to the uncertainty-weighted hybrid prediction. The hybrid uncertainty suppresses unreliable knowledge in the hybrid prediction, leaving only useful information to improve network performance further. Extensive experiments of binary and multi-class segmentation conducted on three MRI datasets demonstrate the effectiveness of the proposed framework. Code is available at https://github.com/ThisGame42/Hybrid-Teacher.
IKD+: Reliable Low Complexity Deep Models For Retinopathy Classification
Mar 04, 2023Shreyas Bhat Brahmavar, Rohit Rajesh, Tirtharaj Dash, Lovekesh Vig, Tanmay Tulsidas Verlekar, Md Mahmudul Hasan, Tariq Khan, Erik Meijering, Ashwin Srinivasan

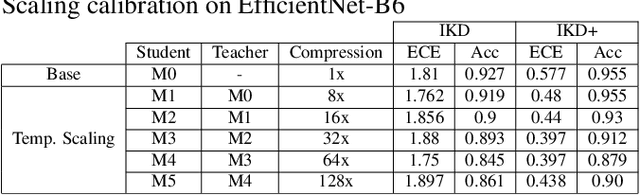
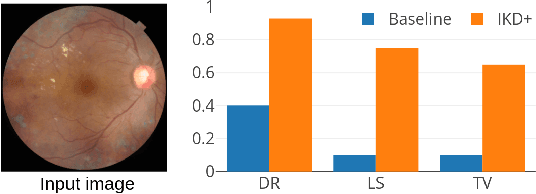
Deep neural network (DNN) models for retinopathy have estimated predictive accuracies in the mid-to-high 90%. However, the following aspects remain unaddressed: State-of-the-art models are complex and require substantial computational infrastructure to train and deploy; The reliability of predictions can vary widely. In this paper, we focus on these aspects and propose a form of iterative knowledge distillation(IKD), called IKD+ that incorporates a tradeoff between size, accuracy and reliability. We investigate the functioning of IKD+ using two widely used techniques for estimating model calibration (Platt-scaling and temperature-scaling), using the best-performing model available, which is an ensemble of EfficientNets with approximately 100M parameters. We demonstrate that IKD+ equipped with temperature-scaling results in models that show up to approximately 500-fold decreases in the number of parameters than the original ensemble without a significant loss in accuracy. In addition, calibration scores (reliability) for the IKD+ models are as good as or better than the base mode
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Annika Reinke, Minu D. Tizabi, Michael Baumgartner, Matthias Eisenmann, Doreen Heckmann-Nötzel, A. Emre Kavur, Tim Rädsch, Carole H. Sudre, Laura Acion, Michela Antonelli, Tal Arbel, Spyridon Bakas, Arriel Benis, Matthew Blaschko, Florian Büttner, M. Jorge Cardoso, Veronika Cheplygina, Jianxu Chen, Evangelia Christodoulou, Beth A. Cimini, Gary S. Collins, Keyvan Farahani, Luciana Ferrer, Adrian Galdran, Bram van Ginneken, Ben Glocker, Patrick Godau, Robert Haase, Daniel A. Hashimoto, Michael M. Hoffman, Merel Huisman, Fabian Isensee, Pierre Jannin, Charles E. Kahn, Dagmar Kainmueller, Bernhard Kainz, Alexandros Karargyris, Alan Karthikesalingam, Hannes Kenngott, Jens Kleesiek, Florian Kofler, Thijs Kooi, Annette Kopp-Schneider, Michal Kozubek, Anna Kreshuk, Tahsin Kurc, Bennett A. Landman, Geert Litjens, Amin Madani, Klaus Maier-Hein, Anne L. Martel, Peter Mattson, Erik Meijering, Bjoern Menze, Karel G. M. Moons, Henning Müller, Brennan Nichyporuk, Felix Nickel, Jens Petersen, Susanne M. Rafelski, Nasir Rajpoot, Mauricio Reyes, Michael A. Riegler, Nicola Rieke, Julio Saez-Rodriguez, Clara I. Sánchez, Shravya Shetty, Maarten van Smeden, Ronald M. Summers, Abdel A. Taha, Aleksei Tiulpin, Sotirios A. Tsaftaris, Ben Van Calster, Gaël Varoquaux, Manuel Wiesenfarth, Ziv R. Yaniv, Paul F. Jäger, Lena Maier-Hein
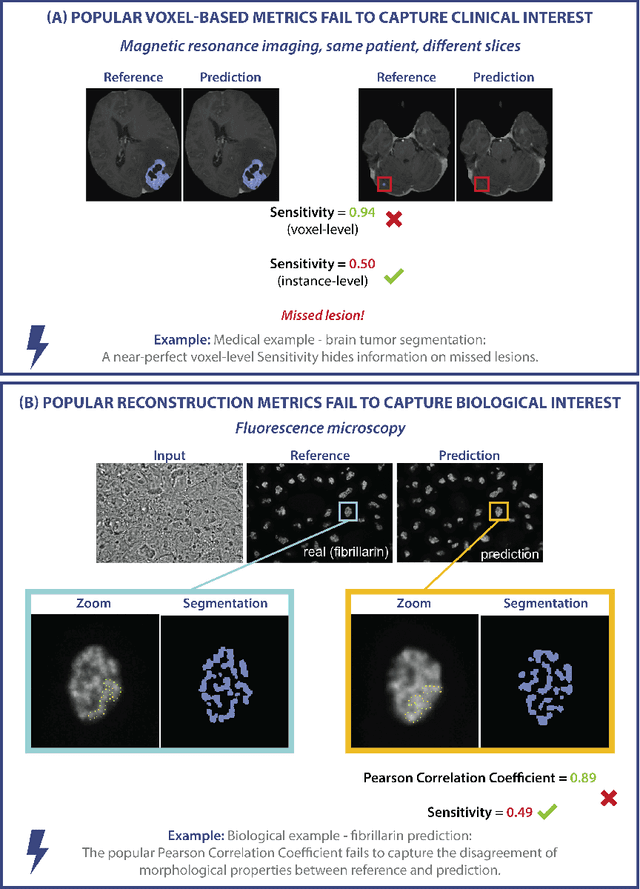
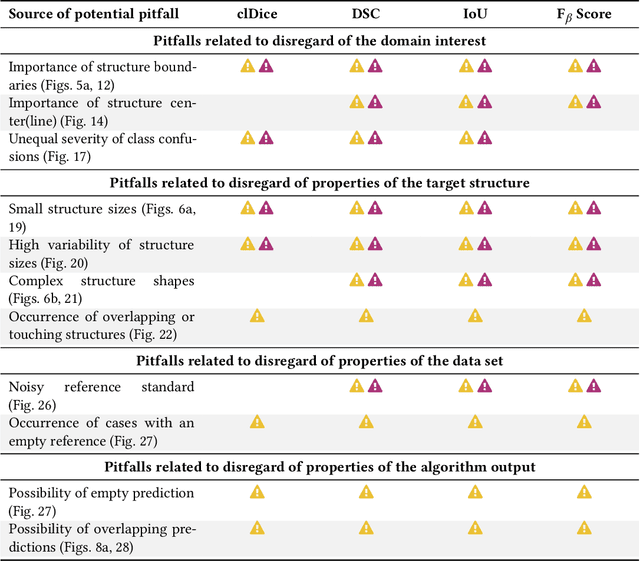
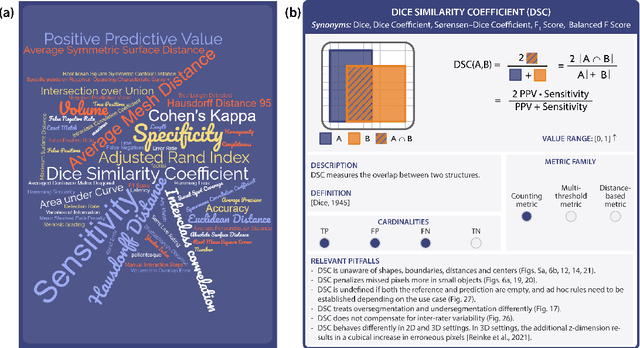
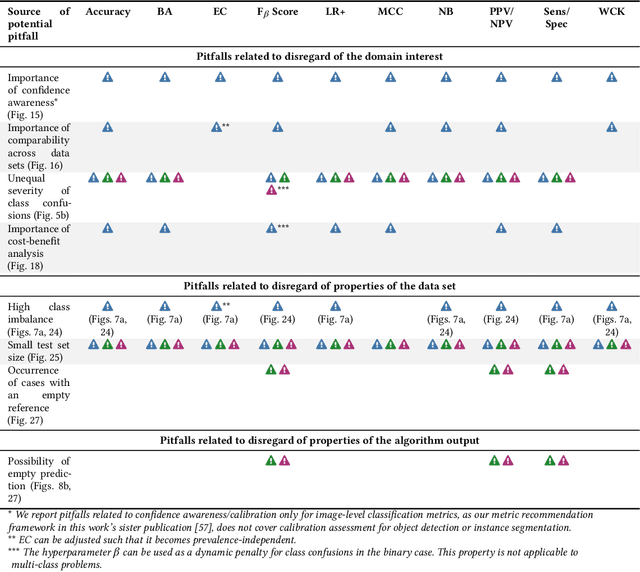
Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Fully Elman Neural Network: A Novel Deep Recurrent Neural Network Optimized by an Improved Harris Hawks Algorithm for Classification of Pulmonary Arterial Wedge Pressure
Jan 16, 2023Masoud Fetanat, Michael Stevens, Pankaj Jain, Christopher Hayward, Erik Meijering, Nigel H. Lovell

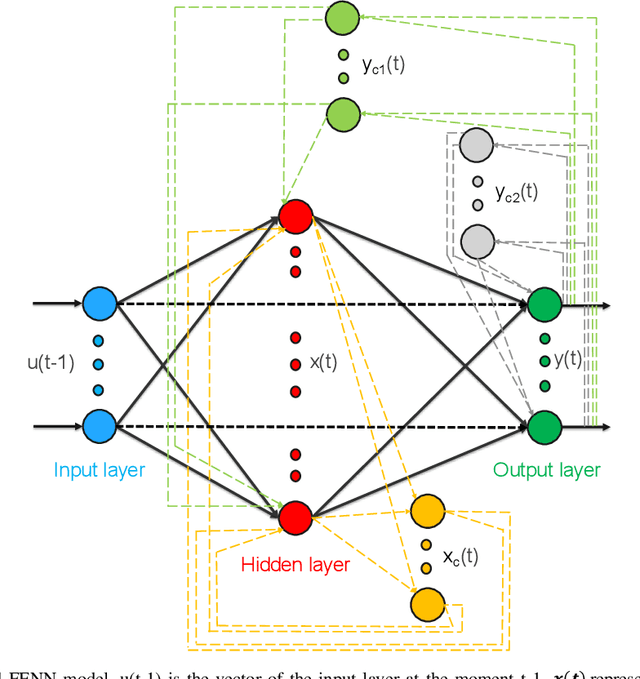
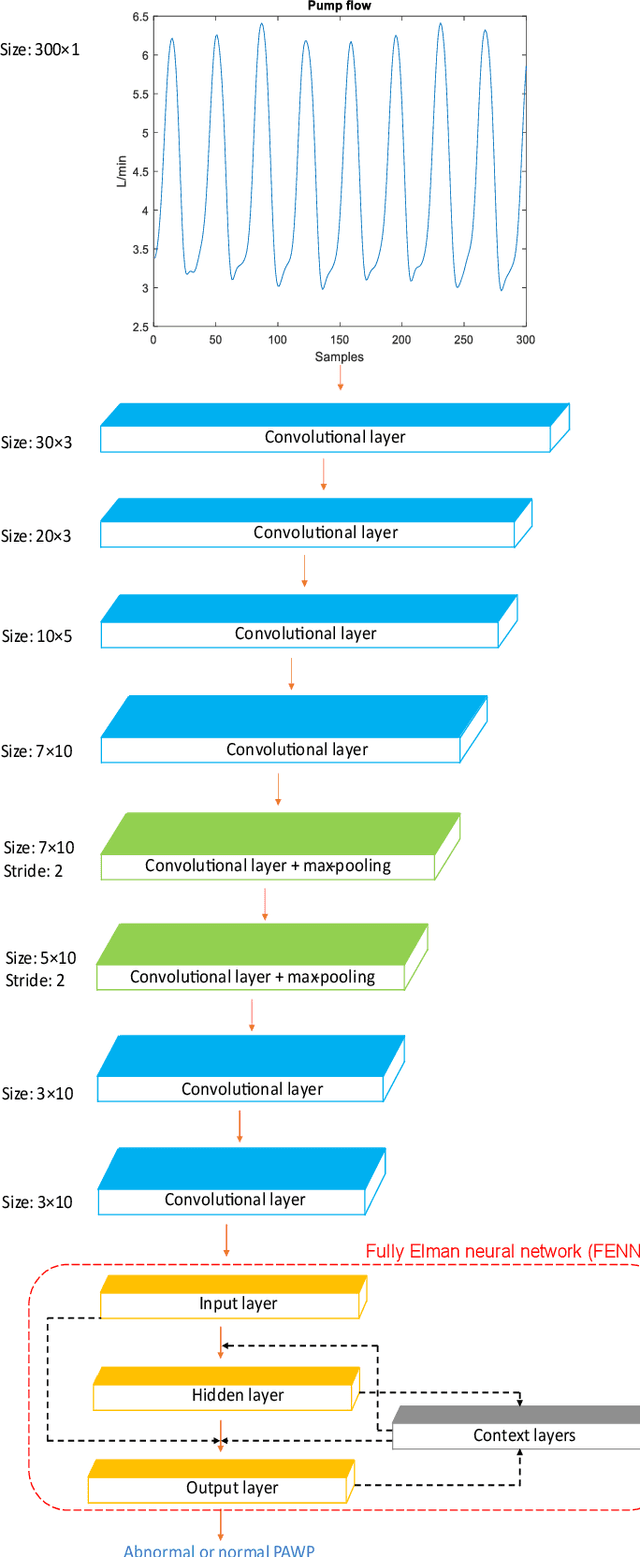
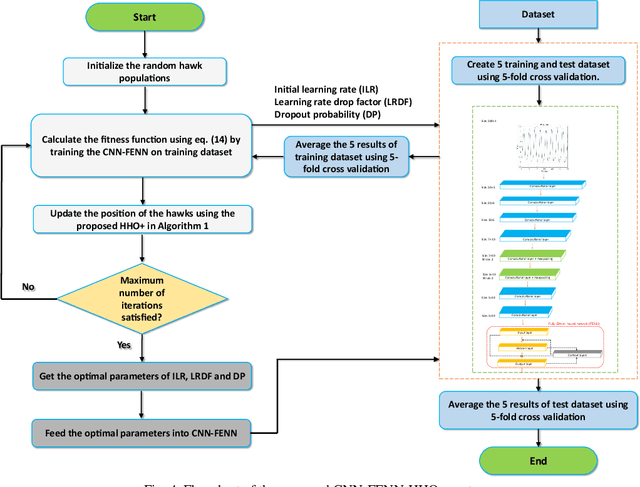
Heart failure (HF) is one of the most prevalent life-threatening cardiovascular diseases in which 6.5 million people are suffering in the USA and more than 23 million worldwide. Mechanical circulatory support of HF patients can be achieved by implanting a left ventricular assist device (LVAD) into HF patients as a bridge to transplant, recovery or destination therapy and can be controlled by measurement of normal and abnormal pulmonary arterial wedge pressure (PAWP). While there are no commercial long-term implantable pressure sensors to measure PAWP, real-time non-invasive estimation of abnormal and normal PAWP becomes vital. In this work, first an improved Harris Hawks optimizer algorithm called HHO+ is presented and tested on 24 unimodal and multimodal benchmark functions. Second, a novel fully Elman neural network (FENN) is proposed to improve the classification performance. Finally, four novel 18-layer deep learning methods of convolutional neural networks (CNNs) with multi-layer perceptron (CNN-MLP), CNN with Elman neural networks (CNN-ENN), CNN with fully Elman neural networks (CNN-FENN), and CNN with fully Elman neural networks optimized by HHO+ algorithm (CNN-FENN-HHO+) for classification of abnormal and normal PAWP using estimated HVAD pump flow were developed and compared. The estimated pump flow was derived by a non-invasive method embedded into the commercial HVAD controller. The proposed methods are evaluated on an imbalanced clinical dataset using 5-fold cross-validation. The proposed CNN-FENN-HHO+ method outperforms the proposed CNN-MLP, CNN-ENN and CNN-FENN methods and improved the classification performance metrics across 5-fold cross-validation. The proposed methods can reduce the likelihood of hazardous events like pulmonary congestion and ventricular suction for HF patients and notify identified abnormal cases to the hospital, clinician and cardiologist.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge